Research
, Volume: 15( 6) DOI: 10.37532/tsbt.2019.15(6).197Proteomic Profile Analysis of Tomato (Solanum lycopersicum L.) Roots Under Salinity Stress
- *Correspondence:
- Amir Jalili
Department of Biology, Urmia University, Urmia, Iran
E-Mail: amir.jalile@yahoo.com
Received: October 28, 2019; Accepted: December 24, 2019; Published: December 31, 2019
Citation: Jalili A, Abbaspor N, Pourakbar L, et al. Proteomic Profile Analysis of Tomato (Solanum lycopersicum L.) Roots under Salinity Stress. Biotechnol Ind J. 2019;15(6):197.
Abstract
Salinity is a major constraint to crop productivity. This study aimed at a better understanding of the molecular adaptation mechanisms of salinity stress was carried out on tomato (Solanum lycopersicum L.) Early Urbana Vf cultivar, using proteomic analysis. Seedlings of tomato were cultivated in the hydroponic culture system. The salt stress (NaCl) was applied (0 mM and 90 mM) and sustained for two weeks. Protein content was extracted from the seedlings roots of both non-saline control and salt-stressed plants, and separated by two-dimensional gel electrophoresis. 22 protein spots were identified and classified into seven categories. Obtained results from two-dimensional gel electrophoresis and proteome analysis indicated that some proteins exhibited expression variations strictly related to salt stress which could be related to genotype tolerance or biochemical Compatibility. The proteins identified with up-regulation belonged to the Heat-shock proteins, ATP synthase, Carbohydrate metabolism, Transcription/translation, Detoxification enzymes, and 4 unidentified protein categories. Furthermore, expression pattern analysis of the expressed proteins under salt stress is an important step in identifying mechanisms of stress resistance in different plants including tomatoes.
Keywords
Tomato (Solanum lycopersicum) cultivars; Root; Two-dimensional gel electrophoresis; Proteomic analysis; Salt stress
Introduction
Salinity is one of the most important abiotic stresses in crop agriculture worldwide. Thirty product types supply 90% of our food, most of which display severe yield losses under salinity stress. Increasing agricultural performance at a time of global warming and increasing population is urgent and should, aside from improving saline soils, include attempts to increase crop tolerance to salt stress [1]. Excess salt in soil has a negative effect on plant growth. Twenty present of the world's agricultural region and half of the world's irrigated lands are affected by soil salinity [2].
The destructive effects of salt on plants are the result of both; water scarcity due to the relatively high solute concentration in the soil and a specific Cl− and Na+ stress. Based on the transcription profiling pattern, it has been suggested that soil salinity causes a number of changes in basic photosynthesis, photorespiration, amino acid and carbohydrate synthesis [3]. Most crops tolerate salinity to the threshold level, above which yields decrease as salinity increases. The development of salt-tolerant plants allows better use of saline soils and saline water [4]. To counteract salt stress, plants have several signaling and metabolic processes at the cellular and organ levels. However, our knowledge of these mechanisms is not sufficient due to the complexity and extent of salt stress, which has both an osmotic and an ionic component [5]. To date, the mechanism of plant sensitivity to salinity remains elusive, mainly because it is controlled by multiple genes that affect different aspects of plant growth and development. In general, plants exhibit a dual response to salt stress, entailing early and late responses. The former is related to osmotic stress resulting from the negative water potential of saline soil. The latter is due to Na+ accumulation in leaf tissues [6]. Only plants with modified adaptive mechanisms can avoid the adverse effect of salinity [7]. Salt resistance in plants is generally an inherent ability of the plants for resistance to the undesirable effects of high concentrations of salt in the rhizosphere or in the leaves without significant negative outcomes. Growth rate maintenance, nutrient preservation, preventing the effects of ion toxicity, and stimulation of metabolism changes that improve water balance are probably the most common characteristics of salt-tolerant plants [4]. Proteomics as a new and effective technique offers a new platform for the study of complex biological functions and can serve as a key tool for revealing the interactions between salinity stress and plant species [8]. At present, the diversity of plant proteomes under salinity stress has been studied in different plants [9], among others in Arabidopsis [10], rice [11], wheat [12] and soybean [13]. Salt-stress due to the greater use of non-potable water for irrigation imposes major restrictions on plant growth in saline soils. Soil salinity may cause osmotic and ionic stresses leading to physiological, biochemical and morphological damages to plants [14]. Extensive research had been done to study gene responses under salinity treatment [15] and identify differentially expressed proteins using mass spectrometry [16], but the molecular aspects of how plants can tolerate salinity have not been fully elucidated. Analyzing the mode of expression of salt-inducible genes is one way of revealing the molecular and biochemical mechanisms of salt-tolerance in higher plants [17].
Tomatoes, one of the most important and most extensive crops in the world, are classified as relatively salt-tolerant. Salinity reduces tomato yield [18], but improves fruit quality features, like soluble solids, acid values, and fruit color [4]. Accumulation of Na and Cl is the principal salinity problem. It has been found that high concentrations of salt (Na and Cl) in the plant leaves reach the toxic level in susceptible genotypes much faster than salt-tolerant genotypes. This is attributed to the ability of the roots to omit the (Na and Cl) from the xylem sap flowing to the shoot. Accumulation ratio of Na and/or Cl in the shoot determines genotypic differences in salt-tolerant cultivars [19]. The potential of different genotypes to tolerate increased salt levels were found to be due to longer root, low Na+ accumulation, and greater K+/Na+ quotient [20]. There are many differences in tolerance of tomato varieties to salinity. Specific differences in salt tolerance were acquired by fresh market, cultivated tomatoes [21]. Osmotic adjustment is an adaptation mechanism involved in salt-tolerance, which permits the maintenance of turgor pressure in water scarcity conditions, as it can counteract the effects of the rapid reduction of leaf water potential [22]. Previous studies in the proteomic analysis [23] developed several strategies to adapt to the salinity stress in tomato roots, including support for glycolysis, tricarboxylic acid cycle, photosynthetic system, ATP biosynthesis, amino acid metabolism, defense-related proteins, and detoxification. Ahsan et al. [24] examined the variation of the proteome in tomato leaves subjected to waterlogging stress and revealed that the expression of proteins related to stress defense mechanisms and energy metabolism were increased, while protein biosynthesis-related enzymes and photosynthesis were strictly decreased. tomato (Solanum lycopersicum L.) is one of the most valuable and widespread crops worldwide and has an important place in the human daily diet. Tomato is an old model system for the study of plant genetics, physiology and morphology [3]. The purpose of this study was to identify and evaluate changes in root protein, due to being under salinity stress, in tomato plants (Solanum lycopersicum L.) by comparative proteomic analysis between plants under stress and control samples.
Materials and Methods
Plant growth and stress conditions
Tomato Seeds (Solanum lycopersicum, Early Urbana Vf cultivar), that has already been reported as salt-tolerant was germinated in a growth chamber in darkness at 25 ± 2°C on Petri dishes. Seedlings growth continued until the radicle initiation step between two layers of wet filter paper. Uniformly germinated Seedlings were transferred to a hydroponic medium containing quarter-strength Hoagland's solution. The seedlings were grown under a 16 h Lighting photoperiod. After 25 days (in 5-7 leaf stage) seedlings were divided into two categories containing different solutions (A-B) and maintained for 14 days. Solution A was a quarter-strength Hoagland solution and served as the control treatment. Solution B contained quarter-strength Hoagland and 90 mM sodium chloride.
Protein extraction
Protein content of the tomato roots was extracted by phenol according to Chen et al. [25]. To measure protein, the Standard Procedure for Microtiter-Plates based on the manual for the Sigma protein assay was used.
Two-dimensional gel electrophoresis (2-DE)
Two-dimensional gel electrophoresis was performed using immobilized pH gradients according to the manufacturer’s directions. For analytical and preparative gels, the 13 cm IPG strips (pH 4-7) (Amersham Pharmacia Biotech, UK) were rehydrated overnight with 250 μl of rehydration stock solution (9 M urea, 3% (w/v) CHAPS, 0.5% (v/v) Triton X-100, 2% (lv/v) IPG buffer, and 0.002% (w/v) bromophenol blue), containing 60 μg of protein, at room temperature. Immunoelectrophoresis (IEF) was conducted at 18°C with a Pharmacia Multiphor II. The running condition was: 300 V for 15 min, followed by 500 V for 15 min, 1000 V for 15 min, 1500 V for 15 min, 2000 V for 15 min, 2500 V for 15 min, 3000 V for 15 min, and finally 3500 V for 5 h. The focused strips were equilibrated twice for 15 min in 10 ml of equilibration solution. The first equilibration was performed in a solution containing 50 mM TRIS-HCl, pH 8.8, 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 0.002% bromophenol blue, and 2 mM tributylphosphine (TBP), and the second equilibration was the same except that TBP was replaced by 2.5% (w/v) iodoacetamide. The second dimension was carried out by SDS-PAGE in a vertical slab of 12.5% acrylamide using a SE 600 Series Vertical Slab Gel Unit. Preparative gels were stained with colloidal Coomassie brilliant blue G-250 [4]. Silver-staining was carried out on colloidal Coomassie-staining [26]. In the silver-staining, the sensitization stage was removed, and the (EDTA) phase was replaced with (1% acetic acid). The protein spots in analytical gels were observed by silver-staining [27]. Three independent replicates were performed to confirm all results.
Image analysis
The gel matching was performed to determine the protein by (Z3) and (Delta 2D) software, and available spot pairs were affirmed optically. The frequency of each protein blot was estimated by percentage (% volume). Only those proteins with significant changes were considered.
Mass spectrometry
Modified spots stained by colloidal Coomassie blue (G-250) were cut and in-gel proteolyzed by trypsin enzyme. The resulting peptides were resolved by reversed-phase chromatography (RP-HPLC) and micro sprayer directly into the ESI-trap mass spectrometer. The analysis of protein spots was repeated twice. The data were compared with simulated-proteolysis and fragmentation of previously known peptides in the National Center for Biotechnology Information (NCBI) database.
Results and Discussion
Evaluation of protein profile to salt-stress
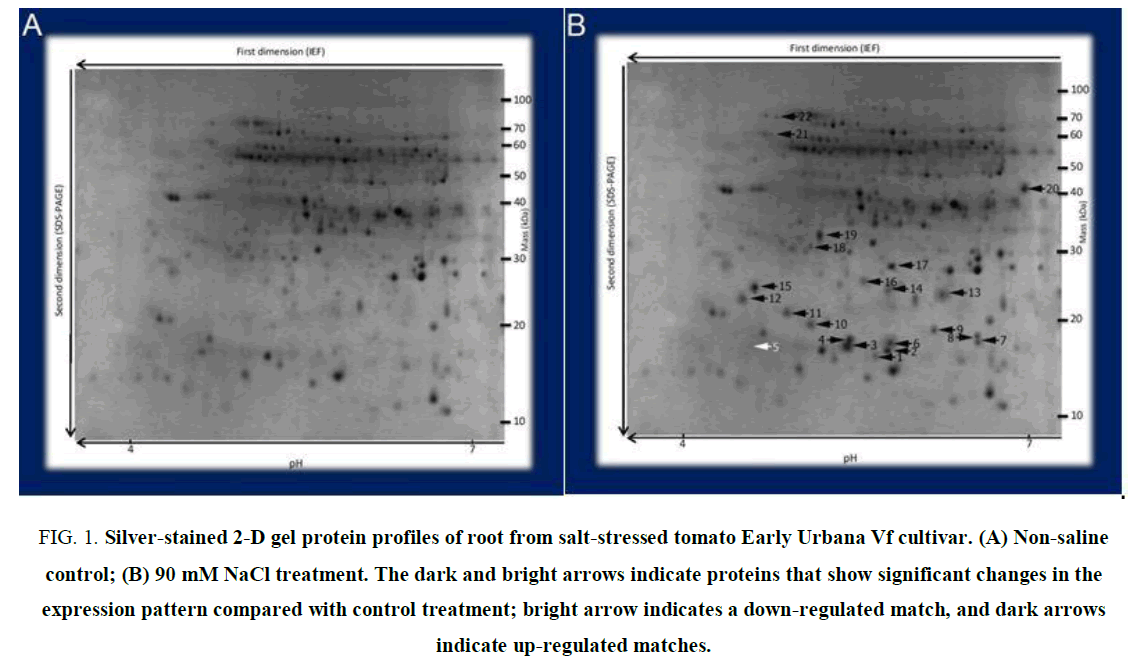
For this purpose protein spots whose levels were altered by salt, stresses were recognized and purified from 2D-PAGE gels. After silver-staining, more than 230 tomato root protein spots were identified and evaluated by digital image analysis. 112 spots gave reproducible staining patterns for all samples Judgmentable by eye and by using (Z3) software. Exposure of Early Urbana Vf cultivar seedlings to 90 mM NaCl for 14 days, revealed 22 root spots (FIG. 1B) that have changed dramatically as compared with control treatments (FIG. 1A). 21 spots were up-regulated, and only spot number 5 was down-regulated (FIG. 1B).
Figure 1. Silver-stained 2-D gel protein profiles of root from salt-stressed tomato Early Urbana Vf cultivar. (A) Non-saline control; (B) 90 mM NaCl treatment. The dark and bright arrows indicate proteins that show significant changes in the expression pattern compared with control treatment; bright arrow indicates a down-regulated match, and dark arrows indicate up-regulated matches.
Classification of identified proteins
All protein spots exhibiting different expression patterns affected by salinity stress were analyzed by (LC-ESI-MS) on a (DECA/LCQ) and recognized by Pep-Miner and request software against the NCBI database (March 2019) of all kinds of herbs. 18 proteins and their Structural information were identified from gene bank studies (TABLE 1). Functionally, 2 proteins were related to carbohydrate metabolism-associated proteins (one 268 amino acid peptide with a weight of 19.0 kDa and one 381 amino acid peptide with a weight of 42.0 kDa), 2 proteins were related to photosynthetic metabolism (one 258 amino acid peptide with a weight of 20.4 kDa and one 332 amino acid peptide with a weight of 33.1 kDa), 3 proteins were associated with detoxifying enzymes (one 185 amino acid peptide with weight of 20.3 kDa and two 250 amino acid peptides with weights of 24.3 and 28.3 kDa), 1 ATP synthase protein (561 amino acid peptide with weight of 18 kDa), 2 proteins were related to transcription and translation functions (one 471 amino acid peptide with weight of 19.3 kDa and one 186 amino acid peptide with weight of 26.2 kDa) and eight proteins were heat-shock proteins (Five 154 amino acid peptides with weights of 16.7, 17.7, 17.9, 18.3 and 18.2 kDa, one 210 amino acid peptide with weight of 21.5 kDa, one 581 amino acid peptide with weight of 62.1 kDa and one 698 amino acid peptide with weight of 71.7 kDa). Other spots, including spots 12, 13, 15 and 19 remained unidentified.
| Spot No. | Observed mol. wt [kDa]/pI | Predicted mol. wt [kDa]/pI | GI number | Classification | Size | Identification |
|---|---|---|---|---|---|---|
| 1 | 16.7/5.7 | 17.8/5.6 | 4836471 | Heat-shock proteins | 154 aa protein sequence | 17.8 kDa class I small heat shock protein (S. lycopersicum) |
| 2 | 17.7/5.8 | 17.8/5.8 | 4836469 | Heat-shock proteins | 154 aa protein sequence | 17.7 kDa class I small heat shock protein (S. lycopersicum) |
| 3 | 17.9/5.6 | 17.7/5.8 | 3341464 | Heat-shock proteins | 154 aa protein sequence | Hsp20.1 protein (Lycopersicon peruvianum) |
| 4 | 18.3/5.6 | 17.6/5.2 | 3336892 | Heat-shock proteins | 154 aa protein sequence | Hsp20.0 protein (L. peruvianum) |
| 5 | 18.0/4.6 | 59.9/5.8 | 3676294 | ATP synthase | 561 aa protein sequence | Mitochondrial ATPase beta subunit (Nicotiana sylvestris) |
| 6 | 18.2/5.8 | 17.6/5.8 | 4836473 | Heat-shock proteins | 154 aa protein sequence | 17.6 kDa class I small heat shock protein (S. lycopersicum) |
| 7 | 19.0/6.4 | 28.9/5.7 | 31088230 | Carbohydrate metabolism | 268 aa protein sequence | Glyceraldehyde 3-phosphate dehydrogenase (S. lycopersicum) |
| 8 | 19.3/6.3 | 51.4/5.5 | 1149571 | Transcription/translation | 471 aa protein sequence | Mitochondrial elongation factor Tu (Arabidopsis thaliana) |
| 9 | 20.3/6.2 | 21.3/6.0 | 77744859 | Detoxification enzymes | 185 aa protein sequence | Temperature-induced lipocalin (S. lycopersicum) |
| 10 | 20.4/5.3 | 27.8/8.3 | 19317 | Photosynthetic metabolism | 258 aa protein sequence | Photosystem II 23 kDa protein (S. lycopersicum) |
| 11 | 21.5/5.2 | 23.8/6.5 | 3492854 | Heat-shock proteins | 210 aa protein sequence | Mitochondrial small heat shock protein (Solanum lycopersicum) |
| 12 | 21.9/4.5 | No match | No match | - | - | No match |
| 13 | 23.0/6.2 | No match | No match | - | - | No match |
| 14 | 23.2/5.7 | 19.6/6.3 | 20018 | Transcription/translation | 186 aa protein sequence | Ribosomal protein L12-1 (Nicotiana tabacum) |
| 15 | 23.8/4.7 | No match | No match | - | - | No match |
| 16 | 24.3/5.7 | 28.1/5.7 | 15080913 | Detoxifying enzymes | 250 aa protein sequence | Ferritin (Malus xiaojinensis) |
| 17 | 28.3/5.7 | 27.3/5.7 | 5257554 | Detoxifying enzymes | 250 aa protein sequence | Cytosolic ascorbate peroxidase (Fragaria3ananassa) |
| 18 | 33.1/5.2 | 35.3/5.9 | 809113 | Photosynthetic metabolism | 332 aa protein sequence | 33 kDa precursor protein of oxygen-evolving complex (S. tuberosum) |
| 19 | 36.4/5.4 | No match | No match | - | - | No match |
| 20 | 42.0/6.8 | 42.0/6.6 | 11991527 | Carbohydrate metabolism | 381 aa protein sequence | Mitochondrial formate dehydrogenase precursor (Solanum tuberosum) |
| 21 | 62.1/5.0 | 61.4/5.2 | 3790441 | Heat-shock proteins | 581 aa protein sequence | Chaperonin 60 a subunit (Canavalia lineata) |
| 22 | 71.7/5.1 | 74.1/5.1 | 77554415 | Heat-shock proteins | 698 aa protein sequence | Chaperone protein DnaK (Oryza sativa (japonica cultivar group)) |
TABLE 1. Expression pattern of the proteins under salt stresses.
Discussion
The response of plants to salt stress is very complex but there are probably some mechanisms to report increased levels of ions, either in the external medium or within the symplast. However, how plants sense Na+ or Cl− remains unknown [28]. Salts affect many cellular processes, primarily because they affect the cell's water content [29]. Salt stress affects plants in several ways: Water stress, ion toxicity, nutritional disorders, oxidative stress, alteration of metabolic processes, membrane disorganization, reduction of cell division and expansion, genotoxicity [30]. Together, these effects reduce plant growth, development, and survival. Salinity causes nutrient uptake inhibition and photosynthesis reduction. It also causes the generation of reactive oxygen species and ultimately plant destruction. In order to reduce the harmful effects of salinity stress, plants have several complicated adaptation mechanisms. Accordingly, it is not surprising to see salt stress in tomato plants changes the balance of carbohydrate metabolism-associated proteins, photosynthetic metabolism, detoxifying enzymes, ATP synthase, transcription and translation, HSPs and intracellular biomolecular equivalence. Although it’s possible that there are some other proteins that were not identified in the present study, it is believed that this amount was ignorable. Silver-staining has several limitations. Therefore, in this study, the frequency of all the differential spots was freely measured. This study is included in the MS-based Specification of proteo-bioinformatics. A proteomic difference property between the salt-stressed treatment and the non-saline control is crucial given its expression pattern of the protein. The analysis of isolated and enriched salt-stressed protein responsiveness showed that 22 detectable protein spots can be evaluated in protein profiles. However, 4 spots remained unidentified. In the roots, significant differences can be identified in the expression pattern of proteins, which may explain the effects of salinity on salt-stressed treatment. Most of the differentially expressed proteins are stress-related proteins engaged in cell defense, rescue, and virulence. Stress-related proteins included the detoxifying enzymes, Temperature-induced lipocalin, Ferritin, Cytosolic ascorbate peroxidase and eight HSPs. Oxygen deprivation is known to produce oxidative stress through the production of Reactive Oxygen Species [ROSs] and accordingly activation of ascorbate peroxidase and superoxide dismutase should be vital to conserve the redox status of the cells [4]. Oxidative stress due to salt stress leads to the formation of some hydroxyl radicals from the reactivity between H2O2 and ferrous iron. In plants, lipocalins have been identified as key enzymes of the xanthophyll cycle which have the function of protecting against photo-oxidative destruction [31]. Most HSPs act as molecular chaperones [32] and assist in protein assembly, transport and folding. Overall, it has been suggested that HSPs protect other proteins from denaturation and aggregation following exposure to stress [33]. Therefore, they may play an important role in protecting the cell against salinity stress. Indeed, it has already been reported that chaperones accumulate in the roots of maize after being affected by stress conditions [34,35]. One of the valuable consequence of this research is the recognition of precursor proteins of the oxygen-evolving complex in roots. Overexpression of oxygen-evolving complex in roots may be a reason for its lateral functions under salinity stress. This study indicates that expressions of 21 proteins were significantly increased in Early Urbana Vf roots. It is clear to find that overexpression of these proteins could partly explain this cultivars' salt tolerance.
Conclusion
Our further knowledge of the protein expression pattern, composition and structure of the plants under salinity stress pave the way to a better understanding of its assembly and functions under stress conditions. Definitely, our more knowledge of the molecular events within the cell will be useful in understanding the mechanisms of stress resistance. This research provides novel perceptions into the response of tomato plants in its tolerance to salinity stress and accentuates the strength and efficacy of a proteomic approach in plant biological studies. The proteomic study revealed some general proteome variations linked to salinity stress. This study also indicates that the overexpressed proteins comprise five functional categories and all the desired proteins are preserved n plants under stress. These findings indicate that the overall function and performance of the stress proteins are conserved. The proteins identified in tomato roots are involved in a variety of processes i.e. oxidative stress, energy production, defense processes and while some are likely part of the overall response to stress which helps plants survive in the suboptimal situations. The quantitative and qualitative analysis of the differentially expressed proteins under salt stress is an important step to clarify more salt tolerance mechanisms and the progress of tomato tolerance to salt stress by eliminating some proteins that control the severity of stress or expression of key salt-stress proteins. Salt stress confines plant growth and development, and reduce the crop index of tomato. Conservation of native protein composition and reduction of non-native protein accumulation is essential for survival especially as such stresses lead to protein accumulation causing metabolic-disruption. HSPs (Heat Shock Proteins) play an undeniable role in salt stress resistance. HSPs induction is one of the most important defense mechanisms of plants in stress conditions. There is also a close relationship between HSPs and ROS amounts, indicating that plants benefit from ROS toxicity and can use ROSs as a stimulator to induce heat shock proteins for better compatibility by activating a set of molecules. Therefore, according to the previous findings and the results of the present study HSPs along with a large set of proteins that show increased expression under stress conditions like transcription/translation proteins, detoxification enzymes and carbohydrate metabolism proteins are involved in the plant survival. Many other salt stress-responsive proteins still remain unidentified. However, because of post-transcriptional and post-translational events like glycosylation and phosphorylation, the mRNA level is not correlated with the amount of protein expression, which is related to signaling pathways and metabolic processes under salinity stress situations. More validation of data reported herewith provides a deeper insight into the stress physiology of this valuable crop plant and paves the way for further studies in this field.
Acknowledgments
Many thanks to Dr. Asra Sadeghzade and Dr. Rashid Jamei for their technical assistance.
References
- Zorb C, Geilfus CM, Dietz KJ. Salinity and crop yield. Plant biology. 2019;21:31-38.
- Kaya C, Kirnak H, Higgs D, et al. Supplementary calcium enhances plant growth and fruit yield in strawberry cultivars grown at high (NaCl) salinity. Scientia Horticulturae. 2002;93(1):65-74.
- Manaa A, Mimouni H, Wasti S, et al. Comparative proteomic analysis of tomato (Solanum lycopersicum) leaves under salinity stress. Plant Omics. 2013;6(4):268.
- Chen S, Gollop N, Heuer, B. Proteomic analysis of salt-stressed tomato (Solanum lycopersicum) seedlings: Effect of genotype and exogenous application of glycinebetaine. J Exp Botany. 2009;60(7):2005-19.
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651-81.
- Khalifa NS. Protein expression after NaCl treatment in two tomato cultivars differing in salt tolerance. Acta Biol Cracov Bot. 2012;54(2):79-86.
- Blumwald E. Sodium transport and salt tolerance in plants. Curr Opinion Cell Biol. 2000;12(4):431-34.
- Zhang H, Han B, Wang T, et al. Mechanisms of plant salt response: Insights from proteomics. J Proteome Res. 2011;11(1):49-67.
- Sharma PC, Prashat GR, Kumar A, et al. Physiological and molecular insights into mechanisms for salt tolerance in plants in Innovative Saline Agriculture. Springer. 2016:321-49
- Jiang Y, Yang B, Harris NS, et al. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot. 2007;58(13):3591-607.
- Parker R, Flowers TJ, Moore AL, et al. An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J Experimen Bot. 2006;57(5):1109-18.
- Peng Z, Wang M, Li F, et al. A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Molecular and Cellular Proteomics. 2009;8(12):2676-86.
- Sobhanian H, Motamed N, Jazii FR, et al. Salt stress-induced differential proteome and metabolome response in the shoots of Aeluropus logophiles (Poaceae), a halophyte C4 plant. J Proteome Res. 2010;9(6):2882-97.
- Zhang H, Guo L, Ye J, et al. Responses of leaf stomatal traits and gas exchange process of cherry tomato to NaCl salinity stress. Trans Chinese Soc Agricult Engineer. 2018;34(5):107-13.
- Ashraf M, Harris PJC. Potential biochemical indicators of salinity tolerance in plants. Plant Science. 2004;166(1):3-16.
- Cho WC. Proteomics technologies and challenges. Genome, Proteomics Bioinform. 2007;5(2):77-85.
- Khalifa NS. Protein expression after NaCl treatment in two tomato cultivars differing in salt tolerance. Acta Biol Cracov Bot. 2012;54(2):79-86.
- Huang C, Peng F, You Q, et al. Growth, yield and fruit quality of cherry tomato irrigated with saline water at different developmental stages. Acta Agricult Scandinavica, Soil Plant Sci. 2016;66(4):317-24.
- Kusvuran S, Yasar F, Ellialtioglu S, et al. Utilizing some of screening methods in order to determine of tolerance of salt stress in the melon (Cucumis melo L.). Res J Agriculture Biological Sci. 2007;3(1):40-45.
- Saeed A, Khan AA, Saeed N, et al. Screening and evaluation of tomato germplasm for NaCl tolerance. Acta Agricult Scandinavica Soil Plant Sci. 2010;60(1):69-77.
- Alian A, Altman A, Heuer B. Genotypic difference in salinity and water stress tolerance of fresh market tomato cultivars. Plant Science. 2000;152(1):59-65.
- Hussain WS, Brajendra SN, Haribhushan A, et al. Compatible solute engineering in plants for abiotic stress tolerance-role of glycine betaine. Current Genomics. 2013;14(3):157-65.
- Manaa A, Ben AH, Valot B, et al. Salt and genotype impact on plant physiology and root proteome variations in tomato. J Exp Bot. 2011;62(8):2797-813.
- Ahsan N, Lee DG, Lee SH, et al. A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiologia Plantarum. 2007;131(4):555-70.
- Chen S, Glazer I, Gollo N, et al. Proteomic analysis of the entomopathogenic nematode Steinernema feltiae IS-6 IJs under evaporative and osmotic stresses. Molecular Biochem Parasitol. 2006;145(2):195-204.
- Yan JX, Wait R, Berkelman T. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis: An Int J. 2000;21(17):3666-72.
- Li Y, Li H, Li YF, et al. Evidence for molecular antagonistic mechanism between mercury and selenium in rice (Oryza sativa L.): A combined study using 1, 2-dimensional electrophoresis and SR-XRF techniques. J Trace Elemen Med Biol. 2018;50:435-40.
- Isayenkov SV, Maathuis FJ. Plant salinity stress: Many unanswered questions remain. Frontiers Plant Sci. 2019
- Maathuis FJ, Ahmad I, Patishtan J. Regulation of Na+ fluxes in plants. Frontiers Plant Sci. 2014;5:467.
- Ndimba BK, Chivasa S, Simon WJ, et al. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5(16):4185-96.
- Azam AFE, Rahman MT, Maniruzzaman M, et al. Nutrients content in some vegetables grown in south-central coastal regions of Bangladesh. The Agriculturists. 2018;16(2):43-57.
- Frenette Charron JB, Breton G, Badawi M, et al. Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS letters. 2002;517(1-3):129-32.
- Sumera A, Radhakrishnan A, Baba AA, et al. Beta-thalassemia and molecular chaperones. Blood Cells, Mol Dis. 2015;54(4):348-52.
- Rozenzvieg D, Elmaci C, Samach A, et al. Isolation of four heat shock protein cDNAs from grapefruit peel tissue and characterization of their expression in response to heat and chilling temperature stresses. Physiologia Plantarum. 2004;121(3):421-28.
- Chang WW, Huang L, Shen M, et al. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol. 2000;122(2):295-318.