Original Article
, Volume: 14( 5)Some Characteristics of the Bacteriocin and Cold Shock Protein of the Strain Lactobacillus Plantarum Ul485 Isolated from Chao of Hue Province in Vietnam
- *Correspondence:
- Uyen Quynh Nguyen , Laboratory of Enzyme-Protein, Institute of Microbiology and Biotechnology, Vietnam National University Hanoi, Vietnam, E-Mail: uyennq@vnu.edu.vn
Received: September 10, 2018; Accepted: September 25, 2018; Published: September 30, 2018
Citation: Nguyen UQ, Dong AHT, Nguyen LMT. Some Characteristics of the Bacteriocin and Cold Shock Protein of the Strain Lactobacillus Plantarum Ul485 Isolated from Chao of Hue Province in Vietnam. Biotechnol Ind J. 2018;14(5):171.
Abstract
Lactic Acid Bacteria (LAB) and bacteriocins have been applied in many fields in our life. Among LAB, Lactobacillus plantarum plays some essential roles such as starter cultures or probiotics. In this study, bacteriocin synthesized by the strain L. plantarum UL485 isolated from the fermented “Chao” of Hue province in Vietnam was partly characterized. In addition, cold shock protein (Csp) in the strain and the survival of the bacteria with temperature downshift after freezing were primarily studied. In particular, the effects of pH and temperature on the bacteriocin, containing in the culture supernatant of the strain were evaluated. The bacteriocin was also partly purified via cation exchange chromatography. After cloning, the deduced amino acids of the cold shock protein in the strain were identified. In addition, after freezing, the survival percentage of the bacteria with temperature downshift from 30°C to 8°C at the growing log phase increased in comparison with that of bacteria without temperature downshift.
Keywords
Activity; Bacteriocins; Cloning; Cold shock proteins; Lactic acid bacteria (LAB)
Introduction
Lactic Acid Bacteria (LAB) are considered as GRAS (Generally Recognized as Safe). LAB are capable of synthesizing useful substances such as bacteriocins, organic acids (like lactic acid and acetic acid) and exopolysaccharides [1,2]. Bacteriocins are synthesized by ribosome pathway and they possess antibacterial activities as well as other valuable properties [3]. Many plantaricins have been studied in details [4]. Due to their wide applications, not only LAB but also bacteriocins attract interest of many scientists.
Lactobacillus is a big genus of LAB. Among them, Lactobacillus plantarum plays many essential roles such as starter cultures or probiotics. The survival and capability of producing bacteriocin of L. plantarum are two topics that are of great interest in the food processing industry. Many publications show the functions of cold shock proteins (Csp) in the adaptation process of bacteria to new environmental conditions, especially in temperature downshift [5]. Csp(s) are a family of proteins with molecular weights around 7 kDa and share a high degree of similarity in their sequences. After a rapid temperature downshift, cold shock proteins regulate the adaptation of bacteria to cold stress [6].
In this study, bacteriocin synthesized by the strain L. plantarum UL485 isolated from fermented “Chao” of Hue province in Vietnam was partly characterized. In particular, the bacteriocin containing in the culture supernatant was resistant to temperature (at 100°C, the activity remained at 80.5%) and it worked at quite a broad range of pH (4-10). After purification via cation exchange chromatography, the purity of the bacteriocin increased to 4.89 fold. The gene encoded for cold shock protein (Csp) in the strain was cloned and then the protein was identified as CspC. In addition, the survival of the strain after temperature downshift from 30°C to 8°C for 2, 5 and 8 hours at the growing log phase primitively was evaluated.
Materials and Methods
Bacterial strains
The strain L. plantarum UL485 was isolated from fermented tofu “Chao” of Hue province (Vietnam), and it was identified by 16S rDNA gene sequencing and sugar fermentation pattern analysis by using API CH50 (bioMérieux SA, USA) [7]. The strain was stored at -80°C in MRS medium (Oxoid, Basing-stoke, United Kingdom) with 15% glycerol, and before use, it was propagated in MRS medium at 30°C for 24-48 hours. The indicator strain was Lactobacillus sakei subsp. sakei JCM1157.
All chemicals were purchased from Sigma Aldrich (USA); MRS and LB media were purchased from Oxoid, Basing-stoke, United Kingdom.
Bacteriocin and activity unit (AU) determination
The supernatant, obtained from the culture medium of the strain UL485, was put into wells on the plates containing Lactobacillus sakei subsp. sakei JCM1157. Bacteriocin was determined by antibacterial zone around the wells. Activity unit (AU) of bacteriocin was determined by the following formula [8]:
Activity Unit (AU) = 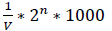
Wherein V is the sample volume (μL), and 2n is the lowest 2-fold dilution of the sample at which bacteriocin activity was not observed.
Effect of temperature and pH on bacteriocin activities
The pH of the samples was adjusted to the values of 5, 6, 7, 8, 9 and 10. After that, the samples were incubated at 30°C for 3 hours and then the bacteriocin activity of the sample at each pH was examined in order to evaluate the effect of pH. Similarly, the temperature (100°C) at different time intervals (0, 5, 15, 30, 45 and 60 minutes) was used to determine the effect of temperature on activities of the bacteriocin.
Bacteriocin purification
Bacteriocin was purified from the cell-free supernatant of the culture medium of L. plantarum UL485 that was grown to the late exponential phase in MRS medium at 30°C. The 6 mL of the sample was loaded on a HiTrap SP FF column of 1 mL on AKTA system (GE Healthcare) with a flow rate of 1 mL/min. After washing with 10 mL of buffer A (sodium acetate 0.02 M, pH=5.5) and then 5 mL of 0.3 M NaCl in buffer A, the bacteriocin was eluted with 5 mL of 0.5 M NaCl in buffer A in fractions of 0.5 mL. The bacteriocin activity of each fraction was detected on agar plates containing the indicator strain. The fractions owning antibacterial activity were pooled to detect total protein by Bradford method [9] and AU [8]. Besides, the total protein and AU of the supernatant, subjected to the chromatography, were determined to evaluate the purity and the yield of the chromatography process.
Electrophoresis
The sample obtained after cation–exchange chromatography was subjected to electrophoresis. Electrophoresis was performed following the Laemmli method in 15% polyacrylamide gel except that the system did not contain both SDS and β-mercaptoethanol [10]. After finishing, the gel was cut into 2 parts. One part containing the sample and low molecular standards (Fermentas) was stained by Coomassie Brilliant Blue R250, while the other part was fixed immediately in a solution of 20% isopropanol and 10% acetic acid for 2 hours and then washed with deionized water for 24 hours. The gel was placed in a petri dish containing about 106 cells of the indicator strain and then the plate was incubated at 30°C for 24-48 hours. After that inhibition zone was evaluated [11].
PCR and cloning
DNA genome extraction and competent cells of Escherichia coli DH5α were performed following the protocol of Sambrook J. et al [12]. The DNA genome of the strain was used as the templates for the amplification of csp gene by using primers (csp1a: 5’ GGTTTAATGTAGACAARGGNTTYGGNTTYAT 3’ and csp1b: 5’ TAGTAGGTACCRTTNGCNGCYTGNGGNCC 3’) with the expected size of 178 bp of PCR products [13,14]. The reaction mixture (50 μL) contained 5 μL of 10X master mix buffer, 50 pmol of each primer, 2.5 U of Taq DNA polymerase, and 1 μL of template DNA. Thermocycles of PCR included 5 minutes of heat shock at 95°C, followed by 30 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 minutes and a final extension at 72°C for 10 minutes. The PCR product was observed by electrophoresis on 1% agarose gel, purified, cloned into pGEM-T vector (Promega) and then transformed into the competent cells of Escherichia coli DH5α. After that the sample was sent to 1st Base company (Singapore) for sequencing. The Clustal Omega, ExPASy tools were used to analyze the nucleotide and amino acid sequences of the csp gene and Csp protein existed in the strain.
Survival assessment
The strain was grown in MRS medium at 30°C for 2.5-4 hours until the OD600 of the culture reached 0.3-0.5. The cultures were divided into some samples: one sample was immediately frozen at -20°C, and the other were cold shocked from 30°C to 8°C for 2, 5 and 8 hours, respectively before they were frozen at -20°C [13,15]. For each sample, before and after freezing, the Colony Forming Units (CFU) by serial diluting in sterile water were counted to give a percentage of viable cells for each sample. All experiments were done in triplicate. The t-test was used for statistics.
Result
Effect of temperature and pH on bacteriocin activity
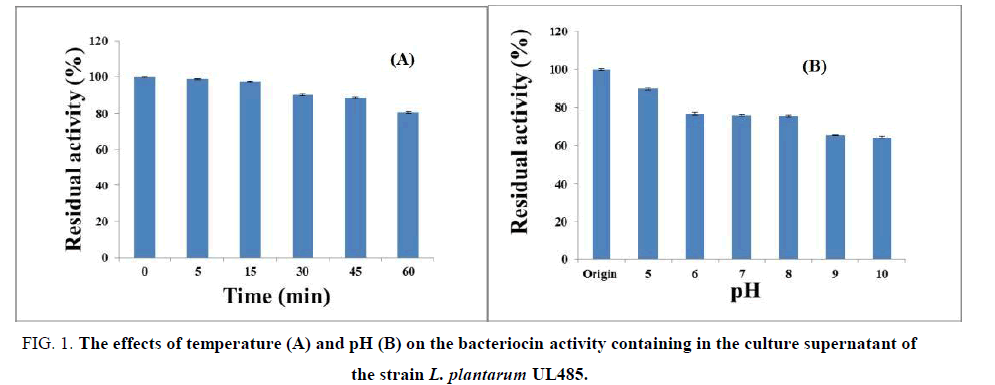
The effects of temperature and pH on the bacteriocin containing in the culture supernatant of the strain UL485 were evaluated (FIG. 1).
Figure 1: The effects of temperature (A) and pH (B) on the bacteriocin activity containing in the culture supernatant of the strain L. plantarum UL485.
The result showed the activity of bacteriocin containing in the culture supernatant of the strain L. plantarum UL485 remained at 80.5% at 100°C and the bacteriocin worked at a pH of 4-10.
Purification of the bacteriocin synthesized by the strain L. plantarum UL485
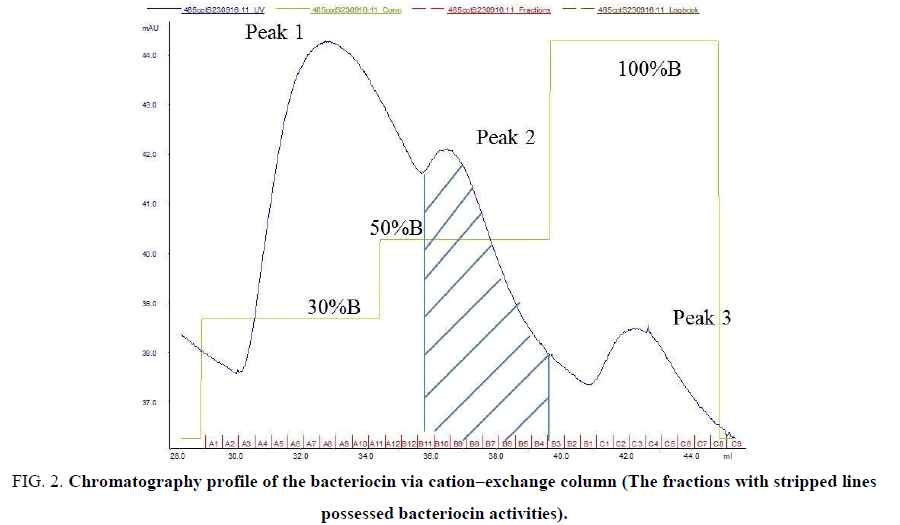
The purification profile and purification result of the bacteriocin synthesized by the strain L. plantarum UL485 via cation–exchange column were presented in FIG. 2 and TABLE 1, respectively.
Figure 2: Chromatography profile of the bacteriocin via cation?exchange column (The fractions with stripped lines possessed bacteriocin activities).
| Sample | Volume (mL) | Total protein (mg)* | Total activity AU/mL** | Specific activity (AU/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture supernatant | 6 | 1.644 | 3200 | 11678.832 | 1 | 100 |
| After chromatography | 4 | 0.028 | 400 | 57142.857 | 4.893 | 8.333 |
* Protein concentration was determined by Bradford method [9]
** Bacteriocin activity (in arbitrary units [AU]) was determined as shown in the method [8]
Table 1: Solubilization index (SI) of PSB isolates isolated from rhizosphere of coffee plant in Pikovskaya?s agar plate, calculated as the ratio of diameter of halo (mm)/diameter of colony(mm).
In order to roughly estimate the molecular mass of bacteriocin, the sample obtained after cation exchange chromatography was subjected to electrophoresis (FIG. 3).
Figure 3: Electrophoresis of the sample obtained after cation exchange chromatography.
The result of electrophoresis showed that the molecular weight of the bacteriocin obtained after chromatography
was a little higher than 10 kDa.
Cold shock protein and survival of the strain UL485 after temperature downshift
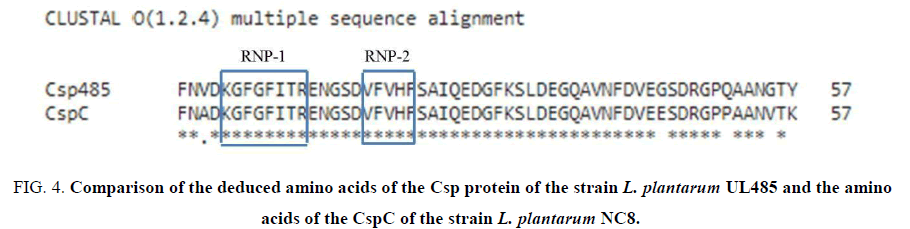
After purifying, the specific PCR product amplified the csp gene of the strain L. plantarum UL485 was cloned into pGEM-T vector (Promega) and its nucleotide sequences were obtained (the result not shown here). The deduced amino acids from these nucleotide sequences showed that the Csp protein contained 2 conserved RNA binding motifs (RNP-1 and RNP-2). Moreover, the RNA binding motifs (RNP-1 and RNP-2) of the Csp proteins of both strain L. plantarum UL485 and strain L. plantarum NC8 were identical. The deduced amino acids of the Csp protein of the strain UL485 and CspC of the strain L. plantarum NC8 were 91.23% identical and 94.74% similar, respectively (FIG. 4).
Figure 4: Comparison of the deduced amino acids of the Csp protein of the strain L. plantarum UL485 and the amino acids of the CspC of the strain L. plantarum NC8.
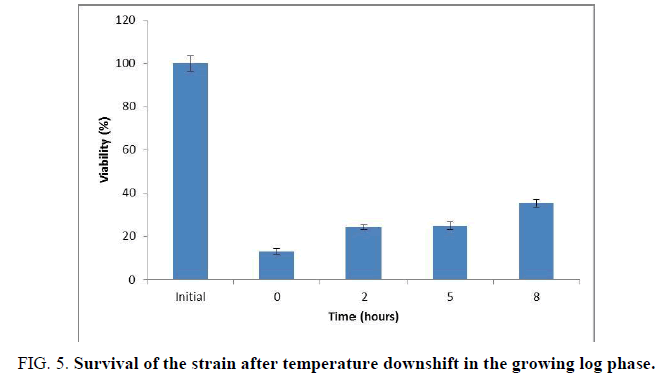
In order to test the function of Csp protein existed in the strain L. plantarum UL485, the survival of the strain after temperature downshift (from 30°C to 8°C) for 2, 5 and 8 hours at its growing log phase was primitively evaluated (FIG. 5).
The result showed that the survival of the strain immediately frozen at -20°C drastically decreased, but it increased gradually in the samples with temperature downshift (from 30°C to 8°C) at the growing log phase for 2, 5 and 8 hours. In particular, the survival percentage of the sample with temperature downshift for 8 hours was 35.41% compared to 13.21% of the sample without temperature downshift.
Discussion and Conclusion
Lactobacillus plantarum is widely used as starters in many variable fermented foods. Therefore its ability to produce useful substances like bacteriocins, lactic acid, exopolysaccharides as well as the ability to survive in different conditions have been of significant interest. In our study, the strain L. plantarum UL485 showed the ability to synthesize bacteriocin and to possess the gene encoded for cold shock protein CspC.
The bacteriocin containing in the culture supernatant of the strain UL485 was stable to temperature (at 100°C, the activity remained at 80.5%), and it worked at quite a broad range of pH (at pH 10, the activity remained at 64.4%). The purification of the bacteriocin of the strain UL485 carried out here was just one step, therefore the purity and the yield of the partly purified bacteriocin were not high. Besides, it is well known that in L. plantarum, plantaricin PlnEF is a two-peptide bacteriocin with the molecular weight of 3.7 and 3.5 kDa respectively. Also, plantaricin PlnA is one peptide pheromone with the molecular weight of 5.4 kDa [4]. The strain UL485 was identified as L. plantarum based on the result of 16S rDNA gene sequencing and sugar fermentation pattern analysis. The genes encoded for PlnEF and PlnA were detected in this strain [7].
In addition, no PCR products were observed when using the specific primers for other plantaricins in the amplification process (the data not shown). Our result of the electrophoresis was consistent with the information that was already known about the strain L. plantarum UL485.
Furthermore, Csp existed in the strain was identified as CspC whose amino acids were 91.23% and 94.74% identical and similar to those of CspC in L. plantarum NC8 [6]. It is well known that Csp proteins play roles in cold adaptation of bacterial cells, but the specific actions and the mechanism via which Csp(s) interacts to fulfill their functions have not been elucidated yet. mRNA of CspC drastically decreased at the stationary growing phase of L. plantarum [16], therefore the role of this protein was tested when the strain’s temperature was downshifted from 30°C to 8°C for 2, 5 and 8 hours at its growing log phase. Our result after freezing showed that the survival of the bacteria increased; however, no significant differences in survival percentage were observed between the bacteria with and without temperature downshift from 30°C to 8°C.
The information reported here for the bacteriocin and the cold shock protein of the strain L. plantarum UL485 can be useful for the application of the strain in our daily life.
Acknowledgment
This research is supported by “Evaluation of typical Lactic Acid Bacteria applicable in foods, pharmaceuticals and animal feeds” grant of the Ministry of Science and Technology.
References
- De Vuyst L, Leroy F. Bacteriocins from lactic acid bacteria: production, purification, and food applications. J. Mol. Microbiol. Biotechnol. 2007;13(4):194-9.
- Khalid K. An overview of lactic acid bacteria. Int. J. Biosci. 2011;1(3):1-3.
- Cotter PD, Hill C, Ross RP. Food microbiology: bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 2005;3(10):777.
- Diep DB, Straume D, Kjos M, et al. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides. 2009;30(8):1562-74.
- Barria C, Malecki M, Arraiano CM. Bacterial adaptation to cold. Microbiology. 2013;159(12):2437-43.
- Wouters JA, Rombouts FM, Kuipers OP, et al. The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst Appl Microbiol. 2000;23(2):165-73.
- Nguyen DA, Luu HT, Hoang TV, et al. Primary study on the bacterial strain UL485 isolated from Chao in Hue province, Vietnam. Agrica. 2018;7(1):82-6.
- Ishibashi N, Seto H, Koga S, et al. Identification of lactococcus-specific bacteriocins produced by lactococcal isolates, and the discovery of a novel bacteriocin, lactococcin Z. Probiotics Antimicrob Proteins. 2015;7(3):222-31.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1-2):248-54.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680.
- Bhunia AK, Johnson MC, Ray B. Direct detection of an antimicrobial peptide ofPediococcus acidilactici in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Ind. Microbiol. 1987;2(5):319-22.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold spring harbor laboratory press; 1989.
- Kim WS, Dunn NW. Identification of a cold shock gene in lactic acid bacteria and the effect of cold shock on cryotolerance. Curr. Microbiol. 1997 Jul 1;35(1):59-63.
- Kim WS, Khunajakr N, Ren J, et al. Conservation of the major cold shock protein in lactic acid bacteria. Curr. Microbiol. 1998 Nov 1;37(5):333-6.
- Derzelle S, Hallet B, Ferain T, et al. Improved adaptation to cold-shock, stationary-phase, and freezing stresses in Lactobacillus plantarum overproducing cold-shock proteins. Appl. Environ. Microbiol. 2003;69(7):4285-90.
- Derzelle S, Hallet B, Francis KP, et al. Changes in cspL, cspP, and cspCmRNA Abundance as a Function of Cold Shock and Growth Phase in Lactobacillus plantarum. J. Bacteriol. 2000;182(18):5105-13.