Original Article
, Volume: 14( 3)Plasmid Construction for Development of a High Throughput System Selection of New Anti-HIV Drugs Derived from Biological Resources Indonesia
- *Correspondence:
- Azzania Fibriani, School of Life Sciences and Technology, Bandung Institute of Technology, Indonesia, Tel: +62-22-2511575; Fax: +62-22-253 41078; E-Mail: afibriani@sith.itb.ac.id
Received: November 09, 2017; Accepted: May 31, 2018; Published: June 05, 2018
Citation: Fibriani A, Feraliana, Steven N, et al. Plasmid Construction for Development of a High Throughput System Selection of New Anti-HIV Drugs Derived from Biological Resources Indonesia. Biotechnol Ind J. 2018;14(3):166.
Abstract
AIDS (Acquired Immunodeficiency Virus) is a disease caused by the HIV virus (Human Immunodeficiency Virus) that attacks the body system. 99% of the causes of AIDS in the world caused by HIV-1. The most effective antiretroviral therapy method currently is HAART (Highly Active Antiretroviral Therapy). One of the therapy components in HAART is an HIV-1 protease inhibitor which has higher genetic barrier than the other HIV treatments. Various studies have been developed to find a novel anti-HIV drug that can be reached by most of the patients in low-limited settings, such as Indonesia. Therefore in this study we constructed a plasmid that can be used for developing a high throughput system as a selection of new anti-HIV drug candidates from Indonesian resources. For plasmid backbone, we used the P00201704939, a constructed plasmid that containing the AraC regulator gene and the GFP reporter gene. The HIV Id protease dimer domain was constructed in plasmid backbone and transformed into host cell Escherichia coli BL21 (DE3) using heat-shock method. Subsequently, transformation results were confirmed by using PCR and Sanger sequencing method. The PCR and sequencing results revealed that the HIV Id homodimer gene was successfully transformed into a backbone plasmid and there was no mutation in the nucleotide and amino acid sequences.
Keywords
HTS, Construct, Protease HIV-1
Introduction
AIDS (Acquired Immunodeficiency Virus) is a disease caused by the HIV virus (Human Immunodeficiency Virus) that attacks the immune system. HIV is a retrovirus with 9,749 nucleotides consisting of two single strands of identical RNA genomes. HIV is divided into two main types namely HIV-1 and HIV-2. Genetically, both types of HIV are similar. However, HIV-2 has a lower virulence level than HIV-1. 99% of the causes of AIDS in the world are caused by HIV-1. In addition, HIV-2 is only present in West Africa. AIDS was first discovered in the United States in 1981 and by 2003 there were more than 70 million people infected with this virus in which 25 million people died [1].
To date, treatment methods have been developed using a combination of anti-HIV drugs to suppress the spread of the HIV virus. The most effective therapy method currently is ART (Highly Active Antiretroviral Therapy). This therapy combines drugs containing HIV protease inhibitors, reverse transcriptase inhibitors and integrase inhibitors [2].
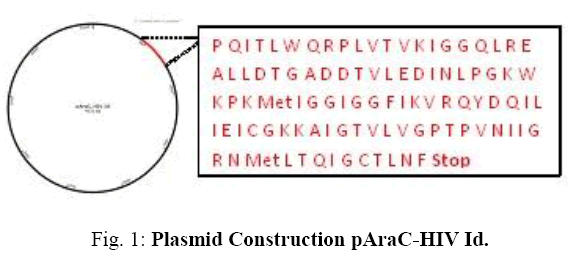
ART therapy causes AIDS-related deaths to decline dramatically and AIDS gradually becomes chronically controlled illness. However, the antiretroviral drugs given to people with AIDS cannot remove the virus from the patient's body. Therefore, drug administration in HIV patients should be done for life. This long duration of treatment may increase the likelihood of non-adherence in taking anti-HIV drugs, leading to rejection of anti-HIV drugs. However, in antiretroviral therapy, there is an HIV-1 protease inhibitor which is one of the most important components in which resistance to the lowest protease inhibitor [2]. Protease is an enzyme that plays an important role in the stage of viral replication. This enzyme breaks down the viral polyprotein precursors so the protein becomes functional and the virus is able to mimic itself. Under natural conditions, dimerization of two HIV-1 protease monomers is an important process for proteolytic protease activity. The formation of HIV-1 protease dimerization occurs in two stages. First, two HIV-1 protease monomers have intermolecular interactions on the active side of each other. Second, there is intermolecular interaction from both ends of each HIV-1 protease monomer [3]. This demonstrates the importance of the dimerization formation stage of two HIV-1 protease monomers so that compounds that inhibit the formation of dimerization of two HIV-1 protease monomers can be good candidates for HIV-1 drugs, as they inhibit HIV replication (Fig. 1).
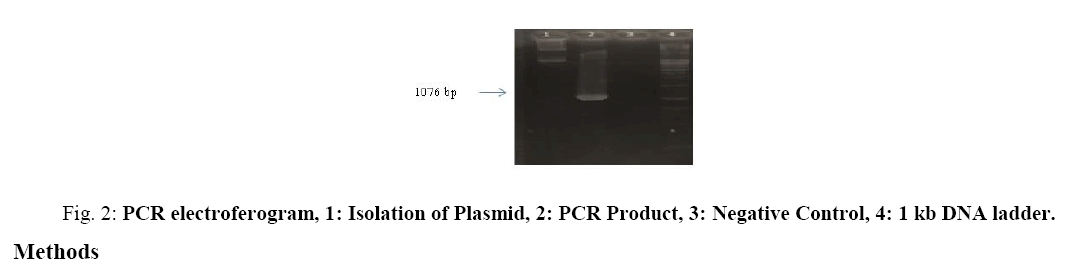
Gervaix et al. [4], has developed method for monitoring HIV infection by cloning a plasmid encoding Green Fluorescence Protein (GFP) to a stable T-cell line (CEM) that driven by the HIV-1 long terminal repeat. In that research, CEM-GFP can be used to monitor HIV infectivity titers and antiretroviral drug susceptibility of syncytium-inducing strains where GFP is nontoxic to cells. Various studies have been developed to find new anti-HIV drugs, but to date these medicines are still not reachable by Indonesians in general. On the other hand, Indonesia is a country rich in natural resources whose potential to become anti-HIV drugs has not been widely studied. Therefore in this study we intend to construct a plasmid that can be used to develop a high throughput system as a selection of new anti-HIV drug candidates derived from Indonesia's biological resources using GFP as marker expression of HIV-1 protease (Fig. 2).
Plasmid construction
The plasmid construction refers to a patent with a P00201704939 number where the PhoR gene of the patent is substituted with a protease-coding gene from HIV (HIV Id). The HIV Id sequence used in this study is the synthesis gene of codon optimization using the software present on the GenScript website. The sequence of HIV Id coded optimization results is then sent to GenScript for synthesis.
Plasmid transformation pRSET-HIV Id
The transformation method used refers to the method of Heat Shock [4] where the host cell used is Escherichia coli BL21 (DE3).
Plasmid isolation
The plasmid isolation was performed using a plasmid isolation kit from Bio Basic [Geneaid].
Plasmid confirmation
PCR method: The transformed plasmid confirmed by PCR method. The PCR profile used for amplification of the AraC-HIV Id homodimer gene was at 95ºC for 3 minutes for predenaturation and denaturation at 95ºC for 30 seconds. Then annealing or primary attachment stage occurred at temperature 56ºC for 30 seconds and continued with elongation stage at temperature 72ºC for 1 minute. This amplification process lasts for 25 cycles. After the 25th cycle, proceed with final elongation at 72ºC for 7 minutes. The PCR result was then electrophoresed. Primary sequences used are forward primer
5'CTGGAAAGGATCCATGGATAATCGGGTACGC'3 and reverse 5'CATAGCACCATGGTTCATACTCCCGCCATTCAG'3.
Sequencing method: Confirmation by sequencing is done by sending samples of plasmid isolation results to the sequencing services company Macrogen Inc. in Seoul, South Korea. Primary sequencing used is a forward primer 5'TAATACGACTCACTATAGG'3 and reverse primer 5 'CATAGCACCATGGTTCATACTCCCGCCATTCAG'3. The sequencing result is then processed by using T-COFFEE software.
Result
Plasmid construction
Based on a patent with P00201704939 number it is explained that this system consists of a gene that can produce a fusion of a protein regulator PhoR with an AraC Escherichia coli repressor and a reporting gene of emerald green fluorescence protein regulated by a fusion protein regulator. In this study, fusion protein regulator PhoR substituted with HIV Id.
PCR confirmation
From PCR electroferogram obtained a single band on the size of 1076 bp. The size of this band corresponds to the size of the AraC-HIV Id homodimer gene band. This suggests that plasmids containing the AraC-HIV Id homodimer gene have been successfully transformed.
Sequencing confirmation
Sequencing analysis was conducted to determine the sequence of nucleotides sequence in a DNA fragment and to know the mutations that occurred after the transformation. Based on Fig. 3 it is known that the value of cons sequence of HIV Id and sequencing results reaches 100 means there is no mutation of nucleotide sequence of the AraC-HIV Id homodimer genes.
Fig 3: Alignment of HIV Id nucleotide sequences with the results of sequencing.
Conclusion
In this study we have successfully constructed the plasmid AraC-HIV Id that might be used for developing a high throughput system as a selection of new anti-HIV drug.
References
- Madigan ML, John MM. Biology of Microorganisms. 11thEd. Pearson Prentice Hall. (2006).
- Lv Z, Chu Y, Wang Y. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV/AIDS (Auckland, NZ). 2015;7:95.
- Hayashi H, Takamune N, Nirasawa T, et al. Dimerization of HIV-1 protease occurs through two steps relating to the mechanism of protease dimerization inhibition by darunavir. Proceedings of the National Academy of Sciences. 2014;111(33):12234-9.
- Hanahan D, Jessee J, Bloom FR. Plasmid transformation of Escherichia coli and other bacteria. In Methods in enzymology, Academic Press. 1991;204:63-113.