Research
, Volume: 15( 4) DOI: 10.37532/0974-7435.2019.15(4).194Phenotypic Characterization and Genotypic Identification of Local Milk Fermented Probiotics
- *Correspondence:
- Bidyut Bandyopyadhyay Department of Biotechnology, Oriental Institute of Science and Technology Vidyasagar University, Dewandighi, Burdwan, West Bengal-713102, Tel: 9733306660; E-mail: bidyut2006@gmail.com
Received: September 13, 2019; Accepted: October 17, 2019; Published: October 28, 2019
Citation: Bandyopyadhyay B, Roy A, Maitra M Phenotypic Characterization and Genotypic Identification of Local Milk Fermented Probiotics. Biotechnol Ind J. 2019;15(4):194
Abstract
Milk fermented product especially curd as a probiotic is a source of lactic acid bacteria with remarkable functional property and has a proved potential therapeutic role in gastrointestinal disease. The objective of this study was to identify genotypic as well as phenotypic characterization of lactic acid bacteria isolated from local curd. The lactic acid bacterial strains which are Gram-positive coccus were tested and proved their ability to inhibit the growth of Vibrio cholera (ATCC 14035). The selected isolate also exhibited bile salt tolerance and antioxidant activity. Thus, among the five isolates, CS3 was proved to be the best potent probiotic. Thus, the optimum pH and temperature of CS3 was determined as pH 7 and 40°C respectively. The morphological characterization of PB 7 was confirmed as coccus in chains through Scanning Electron Microscopy. The 16s rRNA sequencing of the CS3 isolate provides an accession no. MN121704 in NCBI, which is named as Streptococcus thermophilus BURD PB8.
Keywords
Curd; Bile Salt Toleran; Anti-oxidant; 16S rRNA Sequencing; SEM, Streptococcus thermophilus
Introduction
Probiotics have been used as long as fermented milk is been used, but due to their health benefits, it is significant since the last century [1]. Probiotics are essential for the beneficial effect on host nutrition for healthy gastrointestinal function. Probiotics are defined as “live microorganisms when administered in adequate amounts; confer a health benefit on the host” [2]. Thus, probiotic emerges with a beneficial role in medical practice, and the increasing public and scientific interest in the administration of these live micro-organisms to prevent or treat disease came into play [3,4]. Recent studies have suggested that the influence of probiotics extends to widespread inside the body, and somewhat beyond the gut. [5,6] In normal healthy condition, the majority (>90%) of the total cells in the body are present as bacteria in the colon (which is about 1012/gram of large intestinal contents) and develops a protective gut microflora.
But the colonization of different types of bacteria is due to changing food habits and lifestyle. The genus Lactobacillus belongs to the normal and beneficial mucosal microbiota of humans and animals [7] and is important for maintaining the gastrointestinal tract health [8]. Generally, Lactobacilli have been regarded as non-pathogenic and were able to tolerate the acidic condition of the environment, NaCl concentration, and resistance to bile. Lactic acid bacteria (LAB) strains produce bactericidal bioactive agents that are able to control the growth of the pathogens.
The antimicrobial activities of probiotics induce the development of foods for the betterment of the health of consuming health [9]. Different probiotics exhibit very remarkable and noticeable antimicrobial activity against pathogenic bacteria, several suggestions have been proposed for inhibition of pathogenic bacteria, and are also responsible for the production of antimicrobial compounds like; bacteriocins, hydrogen peroxide and organic acids [10-12]. Multiple mechanisms of action for the beneficial effect of probiotics have also been proposed [13-15] along with the ability to adhere to the intestinal mucosa of probiotic microorganisms. The ability of adherance to epithelial cells is an important property for probiotics. Adherence is important for the colonization of probiotics in the intestinal cavity [15].
Ulcerative Colitis in a mouse model can be induced using Dextran Sodium Sulphate (DSS) which is advantageous in comparison to the other chemical induction as it exhibits many symptoms which are similar to those seen in Human Ulcerative Colitis. The symptom of such Ulcerative Colitis includes diarrhea, fecal blood, bodyweight loss, mucosal ulceration, shortening of the large intestine and colon cancer. It is not only possible to induce the acute phase of Ulcerative Colitis but also the chronic one. This model is useful and reliable because of the development and severity of the colitis in individual mice, and the lesions. The microbiological impact on the Ulcerative Colitis of the colon makes it appropriate for the study of probiotic/prebiotic role on the disease. Germinated barley (probiotic) and Clostridium butyricum (Probiotic) is beneficial in the DSS model of Ulcerative Colitis. In another model (IL-10-deficient mice, acetic acid or methotrexate- induced colitis) the developed colitis was treated by Lactobacillus levels or adding exogenous Lactobacilli [16].
Materials and Methods
Further biological evolution of life during ages produced more complex and improved organization of life which correlated to temperature-dependent evolution in the earth [7]. Therefore, it may be concluded that the effect of temperature was unquestionably and contributed to the main dynamic force of phyletic transformation or anagenesis [8]. Collection of samples to isolate desired microorganisms
The study was conducted to isolate and identify the microorganisms from three different samples of Local Curd from Purba Bardhaman district of West Bengal, India. The different curd samples are randomly collected from different local house-made and commercially available at the local market. These samples were collected in clean, sterile, wide-mouthed containers. Immediately after collection, the samples were transferred to the laboratory for microbiological analysis and stored aseptically in low temperature (-20°C) refrigerator to protect from contamination.
Isolation of probiotic bacteria
In our present study, bacteria were isolated from curd samples by using MRS (deMan, Rogosa, and Sharpe) media. 10 grams of each collected sample was diluted with sterilized phosphate-buffered saline (PBS) and transferred to 100 ml of MRS broth which was used for primary isolation of probiotic bacteria. These solutions were added to the MRS broth and streaked on to the MRS agar plates after 6 h of incubation. The plates were aerobically incubated at 37°C for 18-24 h. After incubation, white colonies that formed were selected for single colony isolation.
Identification of the isolates
The isolated colonies formed on the MRS agar plates were phenotypically (Staining and Biochemical Tests) and Genotypically (16s rRNA Sequencing) identified. The identification was performed according to Bergey's manual of determinative of bacteriology.
1. Phenotypic Identification
The Gram staining and Endospore staining were performed with the 10 selected isolates following the techniques of Burkes, 1992 and Cruickshank, 1975. All the isolates were subjected to the biochemical test to identify the isolates. Identification was done on the basis of carbohydrate fermentation test, motility test, catalase, oxidase test, IMViC tests, and Arginine Hydrolase test. [13].
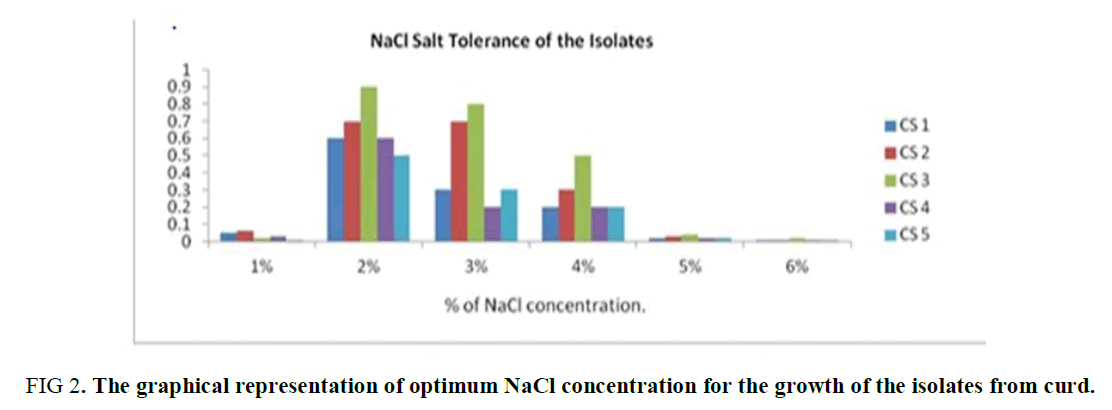
NaCl Tolerance Test: Determination of optimum NaCl concentration of the isolates (CS1-CS5) was monitored by measuring absorbance at 600 nm [17] and NaCl free MRS broth was used as control. The isolates were grown in MRS broth supplemented with different concentrations of NaCl (1-6%) and incubated anaerobically at 37°C for 24 hours of incubation.
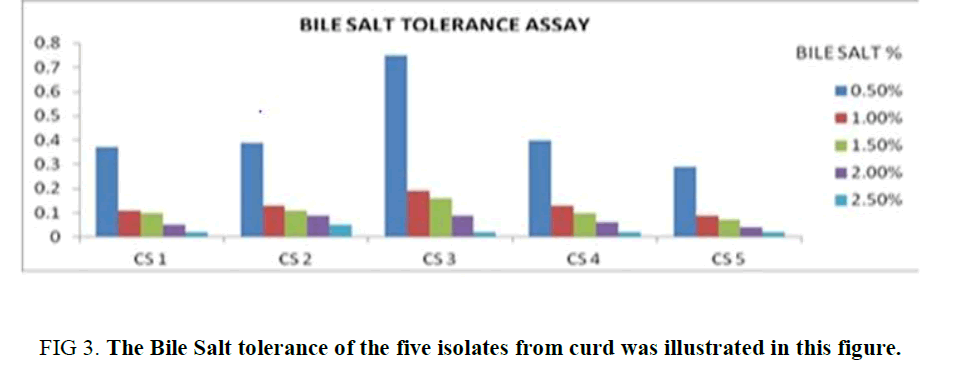
Bile Salt Tolerance Assay: The ability of the bacterial isolates to tolerate bile salts was determined [18]. This bile salt tolerance test was examined by inoculation of all the five isolates (CS1 to CS5) separately into MRS broth tubes containing 0.5%, 1%, 1.5%, 2% and 2.5% bile salts. Bacterial growth was monitored by measuring absorbance at 600 nm after incubation for 24 hours at 37° C.
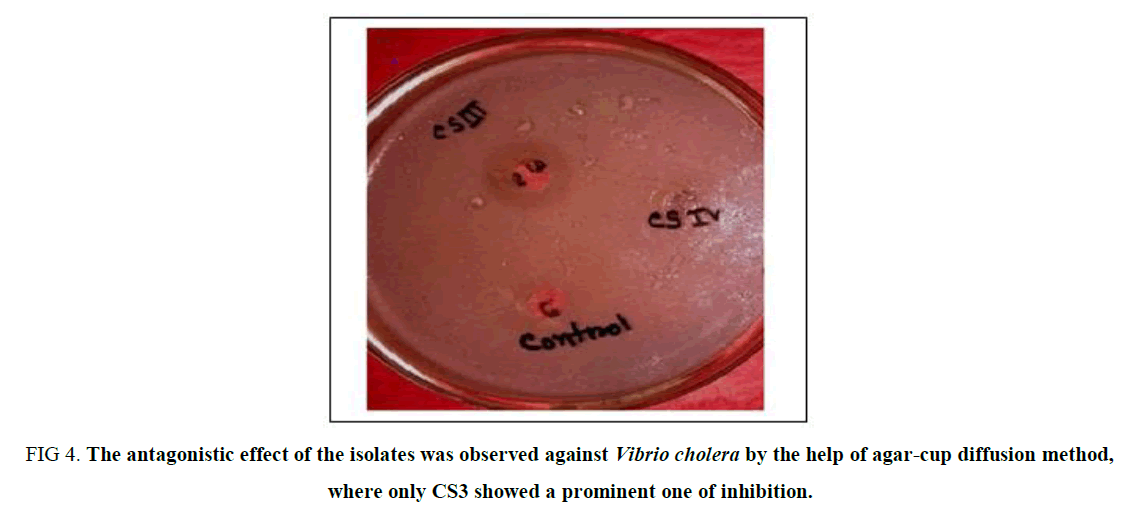
Antagonistic Activity against Pathogens: Antimicrobial activity of selected probiotic isolates (CS1-CS5) against the selected human pathogens were assessed using the agar cup test described by Mami et al., 2012 [19]. An agar well diffusion assay was used, in which sterile cell-free supernatant was placed in 7 mm diameter wells on MullereHinton-agar plates previously seeded with the test pathogen, Vibrio cholera (ATCC 14035). After 24 hours of incubation at 37°C, the diameters of the clear zones of growth inhibition were measured.
Auto-Aggregation of Probiotic Isolates: Aggregation study was examined for all the selected isolates from curd samples (CS1-CS5) on the basis of their sedimentation characteristics. Overnight cultures were harvested by centrifugation at 6000×g for 20 mins, 4°C and washed three times with Phosphate Buffer Saline (pH 7.3) and then eliminated in the same buffer. Then, the mixture was vortex and incubated at 37°C for 4 h. The auto-aggregation percentage was calculated as,
1 – (Atime /Ainitial) × 100
where Atime and Ainitial absorbance measured at 0 hours and 4 hours respectively in 540 nm.
Antioxidant Assay: The scavenging effect of all the five isolates on the free radical DPPH was measured in accordance with the methods used by Lin and Chang [20]. Bacterial samples in MRS broth (1 mL) were mixed with freshly prepared 1 ml DPPH (Sigma-Aldrich) solution (0.2 mM). After shaking the mixture it was left for 30 mins incubation in the dark at room temperature. The control sample only contained deionized water. The scavenged DPPH was then monitored by determining the absorbance at 517 nm using a Spectrophotometer (Systronics, India). The radical scavenging activity was quantified as units/mL (U/mL) by using the following formula [21].
DPPH activity (U/mL) = (ABSC – ABSS)/S × 100
where, ABSC and ABSS are the absorbance of the control and test samples at 517 nm respectively, and S is the volume (mL) of the sample.
Determination of optimal growth at different pH and Temperature: For determining the optimal growth temperature and pH of the isolate CS3, two sets of fresh overnight cultures were taken which was inoculated into i) MRS broth with varying pH ranging from 3, 5, 7, 9 and 11 (SET I) and ii) MRS broth with pH 7 as the other set with varying temperatures at 0°C, 20°C, 40°C, 60°C, 80°C respectively (for SET II). Both the sets of inoculated broths were incubated in anaerobic conditions for 24 hours at 37°C. After 24 hours of incubation growth of the bacteria were measured using a spectrophotometer, reading the optical density at 560 nm against the un-inoculated broth.
Scanning Electron Microscopic Study: As it is difficult to notice small changes in cell morphologies of bacteria under the light microscope, SEM was used in the present investigation to review the changes or any rapture in cell morphology of the five isolates (CS1-CS5) [22]. The bacterial cells from each culture were recovered by centrifugation at 6000 rpm/min and the cells were washed twice with potassium phosphate buffer (50 mM, pH 7.0). Bacterial cells were then fixed in 2.5% glutaraldehyde in potassium phosphate buffer (50 mM, pH 7.0), overnight at 4°C. Then the specimens were washed twice with buffer and dehydrated by ethanol series (v/v) ranging from 30%, 40%, 50%, 60%, 70%, 80%, 90%, and 100% and stored in absolute ethanol. The specimens were then dried to the critical point, coated with gold and inspected with an S-200C scanning electron microscope.
2. Genotypic Identification
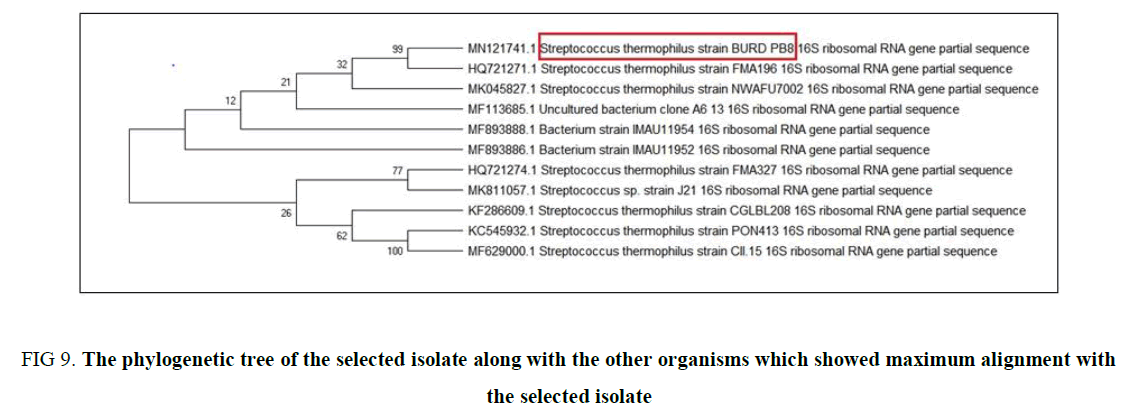
Molecular identification of Lactobacillus spp.: The isolated strain CS1 was identified by 16s rRNA sequencing, after the initial analysis at NCBI. The relevant sequences were downloaded and Phylogenetic analysis was done and a tree was constructed using ClustalW software and showed the Phylogenetic relationship of the isolates. The analysis of the sequence showed a 99% sequence similarity with the sequence reported. The 16s rRNA isolate sequence is deposited in the Gen- Bank with the accession number MN121751.
Results and Discussion
Phenotypic identification by staining procedures
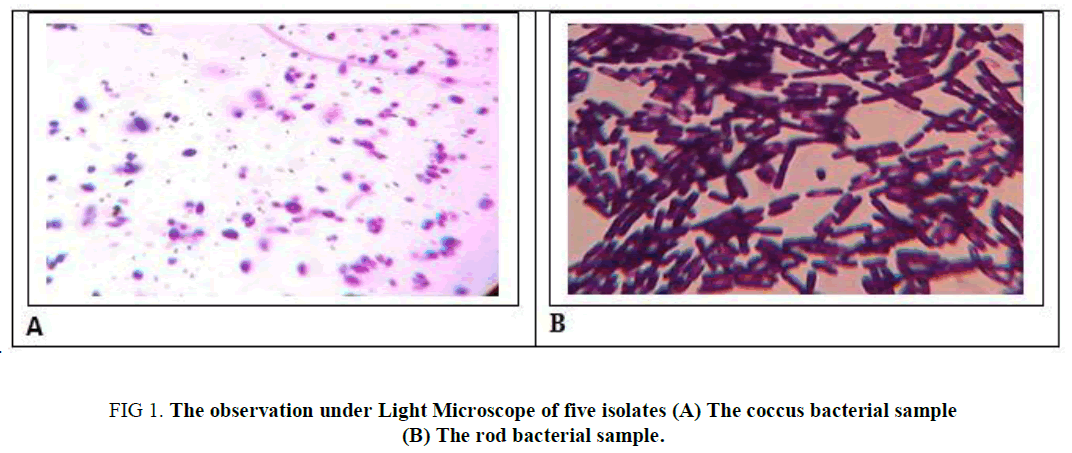
A total of 5 homemade curd samples were collected from the local market of the Dewandighi, Purba Bardhamaan, West Bengal. Five strains were isolated from each source, following the series of subculturing method. And white colonies on the MRS agar plates were observed as rods (Cs, CS4, CS5) as well as coccus ( CS2, CS3) in shape. For further information, they were subjected to endospore staining which gives a negative test as illustrated in Table 1 and observed as in Figure 1.
| Samples | Gram Character | Endospore formation | Morphology |
|---|---|---|---|
| CS 1 | Gram + ve | Non-spore former | Rods in chains or singly |
| CS 2 | Gram + ve | Non-spore former | Coccus in chains or singly |
| CS 3 | Gram + ve | Non-spore former | Coccus in chains or singly |
| CS 4 | Gram + ve | Non-spore former | Rods in chains or singly |
| CS 5 | Gram + ve | Non-spore former | Rods in chains or singly |
TABLE 1. The phenotypic identification of the five isolates with the help of the different staining procedures (such as Gram Staining & Endospore Staining).
Figure 1. The observation under Light Microscope of five isolates (A) The coccus bacterial sample (B) The rod bacterial sample.
NaCl Tolerance Test: From this experiment, it was observed that all the five isolates grow well at 2 to 3 % of NaCl concentration in the media. The growth of the isolates was suppressed in less or more than this range (2-3%). Thus, the optimum range of the NaCl concentration of the isolates is 2 to 3% is observed in Supplementary Table 1 and Figure 2.
Figure 2. The graphical representation of optimum NaCl concentration for the growth of the isolates from curd.
Bile Salt Tolerance Assay: All the isolates showed its tolerance towards bile salt up to 0.5%, but among them, the growth of CS2 and CS3 is significantly high in comparison to the other isolates. The growth of the isolates was observed to be diminished with increasing bile salt concentration in the growth media (1%, 1.5%, 2%, 2.5%). Although in each case the optical density referring to the growth of the CS2 and CS3 was higher in compared to the rest of the isolates in each concentration. (Supplementary Table 2). The observation depicts CS2 and CS3 as a bile salt-tolerant (Figure 3) which is up to 1%.
Antagonistic Effect on Vibrio Cholera: The antagonistic effect of the isolates was observed following the agar cup method against a potent human pathogen Vibrio cholera, and the result was surprisingly unique which gives no zone of inhibition for the isolates except for CS3 (Supplementary Table 3). Thus, it can be concluded that CS3 is the only isolate that can be selected for its probiotic nature due to its anti-microbial nature against Gastrointestinal disease-causing pathogen (Figure 4).
Figure 4. The antagonistic effect of the isolates was observed against Vibrio cholera by the help of agar-cup diffusion method, where only CS3 showed a prominent one of inhibition.
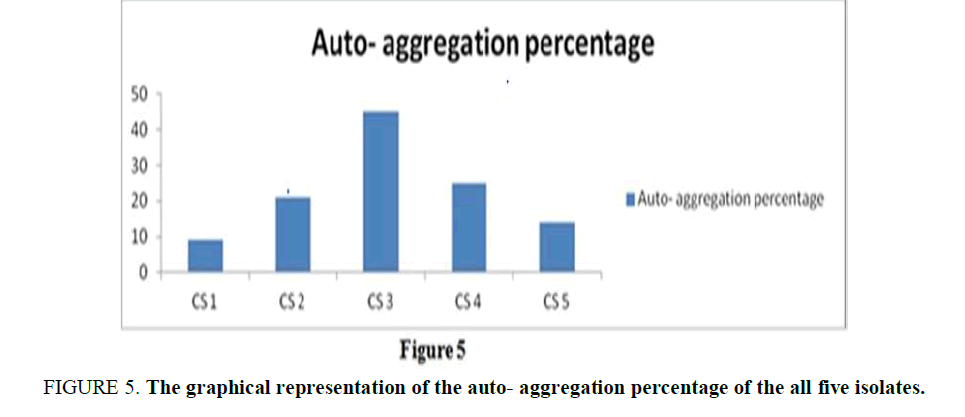
Auto-Aggregation of Curd Samples: The auto-aggregation assay was examined for five isolates on the basis of their sublimate characteristics. The percentage of auto-aggregation for all indicator strains ranged between 9% and 45% (Supplementary Table 4), for 4 hours of incubation. The observation (Figure 5) referred to an obvious conclusion the autoaggregation capacity is highest in CS3, but the rest four isolates also showed a certain percentage of autoaggregation.
Figure 5. The graphical representation of the auto- aggregation percentage of the all five isolates.
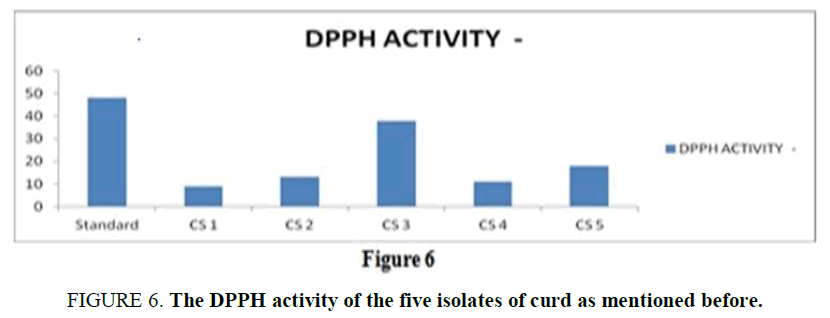
Anti-Oxidant Assay: The anti-oxidant activity is an important property of a potent probiotic, and it is measured in the terms of scavenging activity using the DPPH. For a standard reference, the scavenging activity of ascorbic acid is used as it is a huge source of anti-oxidant known. From the result (Supplementary Table 5), it is observed that all the isolates showed a minimum antioxidant activity whereas, CS3 showed the highest scavenging activity compared with the standard used (Figure 6).
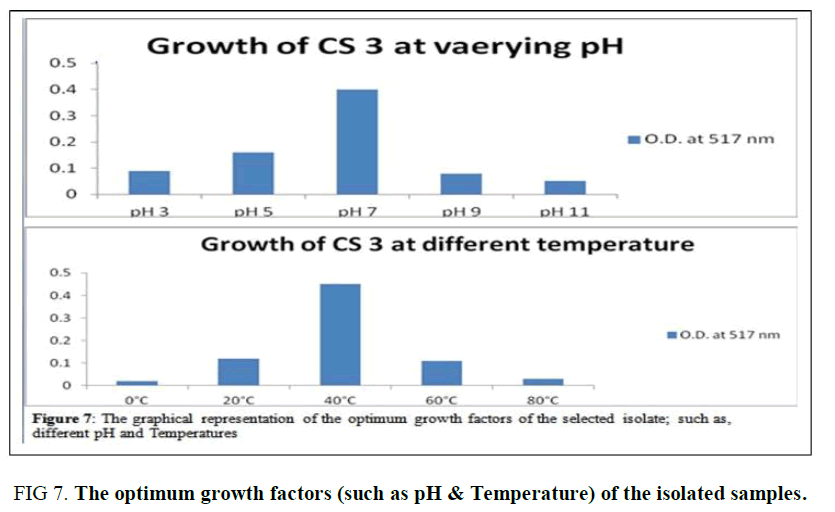
Optimum Growth pH and Temperature of CS3: The selected isolate CS3 is observed to be a potent probiotic among the five isolates as it acts as a good NaCl tolerant, Bile salt-tolerant, Antagonistic effect against Vibrio cholera, Auto- aggregates initiator, and Anti-oxidant agent. Thus, the optimum temperature and pH of CS3 were determined as 40°C and pH 7 as illustrated in (Supplementary Table 6, Figure 7).
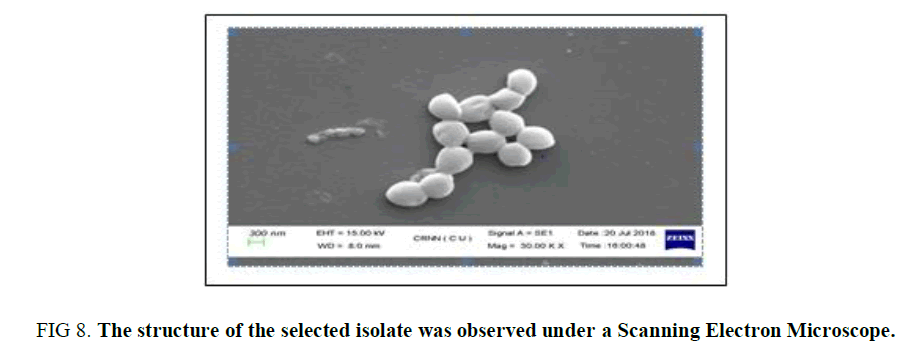
Scanning Electron Microscopic Study: For the detailed observation of the sample CS3, it was observed under Scanning Electron Microscope and was observed (Figure 8) as coccus in shape which is in the chain and in cluster formation. This observation also confirms the texture of the surface of the isolate.
Genotypic Identification and Phylogenetic analysis: The genotypic identification of CS3 was initiated by purification of the isolated DNA and amplified for 16S rRNA sequencing which was performed by KPC Medical College, Kolkata. The partial sequence that we received was initially read and identified its relation with the neighbor bacteria present in the database by forming a phylogenetic tree (Figure 9) using BLASTn. This result leads to a conception that the isolate is not 100% homologous to any of the sequences present in the database. Thus, this partial sequence was submitted to NCBI, which give an accession no. MN121741 and is named as Streptococcus thermophilus BURD PB8.
Figure 9. The phylogenetic tree of the selected isolate along with the other organisms which showed maximum alignment with the selected isolate
Conclusion
The above experiments lead to the conclusion that all the isolates were Gram-positive rods as well as Garam positive coccus and were non-endospore former. Although all the five isolates act a good NaCl tolerant and Bile salt tolerant but the growth capacity is highest in CS3. Furthermore, the CS3 exceptionally a significant-good result in antagonistic effect against Vibrio cholera, auto- aggregation effect, and anti-oxidant activity. The optimum pH and temperature of CS3 were determined as pH 7, 40°C respectively. And thus CS3 supports the growth of the isolates in the GI tract of the human body. These indicate the CS3 to be a potent probiotic, which after 16S rRNA sequencing was assigned as a new strain of Streptococcus thermophilus BURD PB 8 and the accession no. is MN121741.
Acknowledgment
The authors are extremely grateful to the Oriental Institute of Science and Technology, Burdwan, West Bengal for providing laboratory facilities and necessary support for doing the present Research work
Conflict of Interests
Atrayee Roy, Madhumita Mitra, and Bidyut Bandyopadhyay confirm that this article content has no conflicts of interest.
Contributions
Bidyut Bandyopadhyay and Madhumita Mitra helped in designing experiments, analyzing data; and development of methods, material preparation. Experiments were performed by Atrayee Roy and supervised by the other two authors mentioned in prior. Atrayee Roy wrote the manuscript with contributions from Bidyut Bandyopadhyay and Madhumita Mitra. All authors approved the manuscript before submission.
References
- Patel AK, Ahire JJ, Pawar SP, et al. Comparative accounts of probiotic characteristics of Bacillus spp. isolated from food wastes. Food Res Int. 2009;42(4):505-10.
- FAO/WHO evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. In: Food and Agriculture Organization of the United Nations and World Health Organization expert consultation report Rome. 2001.
- Miele E, Pascarella F, Giannetti E, et al. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol. 2009;104(2):437-43.
- Niedzielin K, Kordecki H, Birkenfeld B. A controlled double blind randomized study on the efficacy of Lactobacillus plantarum 299V in patients with irritable bowel syndrome. Eur J Gastroenterol Hepatol. 2001;13(10):1143-47.
- Gareau MG, Wine E, Rodrigues DM, et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60(3):307-17.
- Bravo JA, Forsythe P, Chew MV, et al. Ingestion of Lactobacillus strain regulates emotional behaviour and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA. 2011;108(38):16050-55.
- Lahteinen T, Malinen E, Koort JM, et al. Probiotic properties of Lactobacillus isolates originating from porcine intestine and feces. Anaerobe. 2010;16(3):293-300.
- Gu RX, Yang ZQ, Li ZH, et al. Probiotic properties of lactic acid bacteria isolated from stool samples of longevous people in regions of Hotan, Xinjiang and Bama, Guangxi, China. Anaerobe. 2008;14(6):313-37.
- Eduardo L, Chuayana JR, Carmina VP, et al. Antimicrobial activity of probiotics from milk products Phil. J Microbiol Infect Dis. 2003;32(2):71-4.
- Charlier C, Even S, Gautier M, et al. Acidification is not involved in the early inhibition of Staphylococcus aureus growth by Lactococcus lactis in milk. Int Dairy J. 2008;18(2):197-203.
- Lin WH, Hwang LW, Chen HY, et al. variable counts characteristic evaluation for commercial lactic acid bacteria products. Food Microbiol. 2006;23(1):74-8.
- Hernandez D, Cardell Zarate V. Antimicrobial activity of lactic acid bacteria isolated from Tenerife cheese: initial characterization of plantaricin TF711, a bacteriocin-like substance produced by Lactobacillus plantarum TF711. J Appl Microbiol. 2005;99(1):77-84.
- Ng SC, Hart AL, Kamm MA, et al. Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis. 2009;15(2):300-10.
- Sherman PM, Ossa JC, Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutr Clin Pract 2009; 24(1):10-4.
- Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008;72(4):728-64.
- Okayasu I, Hatakeyama S, Yamada M, et al. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98(3):694-702.
- Quwehand AC, Vesterlund S. Antimicrobial components from lactic acid bacteria. Lactic acid bacteria, microbiological and functional aspects. 2004:375-96.
- Graciela FVD, Maria PT. Food microbiology protocols probiotic properties of Lactobacilli. Totowa: Spencer Humana Press Inc, 2001.
- Mami AZ, Boumehira AR, Hamedi JE, et al. Screening of autochthonous Lactobacillus species from Algerian raw goats' milk for the production of bacteriocin-like compounds against Staphylococcus aureus. Afr J Microbiol Res. 2012;11(20):88-98.
- Lin M, Chang F. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356, US National Library of MedicineNational Institutes of Health. 2000;45(8):1617-22.
- Gazi MR, Yokota M, Tanaka Y, et al. Effects of protozoa on the antioxidant activity in the ruminal fluid and blood plasma of cattle. Anal Chim Acta. 2007;78(1):34-40.
- Raouf T, Marion B, Marielle G, et al. In vitro characterization of aggregation and adhesion properties of viable and heat-killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni. J Med Microbiol. 2013;62(4):637e49.