Short communication
, Volume: 17( 1)From Human Structural Disorders to Basic Physical Chemistry to Protein Dynamics, Folding and Misfolding
- *Correspondence:
- Andrew Smith
Department of Organic Chemistry, Ghent University, Ghent, Belgium
E-mail: smith_andrew@yahoo.com
Received: December 27, 2022, Manuscript No. TSOC-22-84765; Editor assigned: December 29, 2022, PreQC No. TSOC-22-84765 (PQ); Reviewed: January 12, 2023, QC No. TSOC-22-84765; Revised: February 27, 2023, Manuscript No. TSOC-22-84765 (R); Published: March 07, 2023, DOI:10.37532/0974-7516.2023.17(1).004
Citation: Smith A. From Human Structural Disorders to Basic Physical Chemistry to Protein Dynamics, Folding and Misfolding. Org Chem Ind J. 2023;17(1):004.
Abstract
Proteins can move in a variety of ways, including the rotation of the side chains of amino acids and the movement of large domains. Because of the conformational flexibility of proteins, it is believed that they can rapidly and dynamically switch between a varieties of conformational substates. This theory has received support from numerous experimental techniques and computer simulations of protein dynamics. Investigations of the subunit dissociation of oligomeric proteins driven by hydrostatic pressure suggest that the typical times for subunit exchange between oligomers and for interconversion between different conformations may be rather slow (hours or days). Instead of an ensemble of conformational substates that rapidly interconvert, proteins should be seen in this context as a consistently diversified population of various long lived conformers.
Keywords
Protein dynamics; Energy minima; Physicochemical states; Triose phosphate isomerase; Creutzfeldt-Jakob disease
Introduction
Since Annsen’s groundbreaking research, protein collapsing has primarily been understood as a cycle in which a polypeptide chain hunts in conformational space to achieve the alleged “local” compliance compared to the global free energy least under a specific configuration of physicochemical states of the medium. Early on, it was recognized that collapsing did not occur by irregular testing of all open conformations because of the enormous amount of the conformational space accessible for collapsing of even the smallest proteins [1]. All other things being equal, it is currently believed that the conformational search takes place in an energy environment resembling a multi layered channel, the slant of which selectively directs the protein downward toward the energy least. The channel causes a significant increase in collapsing rate (in comparison to the normal rate for an irregular dilusional process) and to some extent, prevents capture in collapsed states (nearby energy minima). Recent findings, however, indicate that favourable to tein collapse to an organically skilled compliance does not always or essentially result in a state that is different from the generally speaking free energy least [2]. The existence of metastable adaptations has occasionally been depicted, suggesting motor control rather than thermodynamic control of the collapse process in some instances, collapsing appears to continue even after reaching one's maximum natural potential (for example a slow strengthening of the protein structure).
Description
Protein dynamics
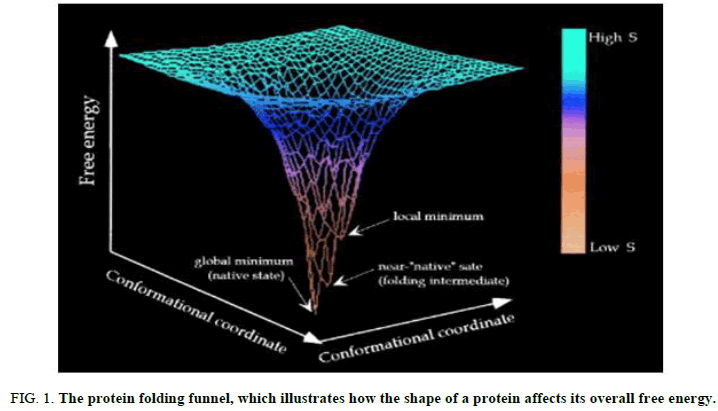
Hierarchical conformational substates: Over the past 20 years, significant knowledge has been gathered about the energetics, kinetics and routes involved in the folding of various different proteins [3]. Efforts have also been made to incorporate experimental findings into conceptual frameworks that could be broadly applicable to describe folding. In this context, the concept that proteins fold in a funnel shaped energy landscape has been introduced, and it appears to be particularly helpful in explaining some of the fundamental problems with folding. However, according to the funnel model, folding ends at a singular, native state that corresponds to the system’s overall free energy minimum (Figure 1) [4]. Given the aforementioned factors, it would appear more acceptable to take into account the potential for several conformers to exist at the funnel’s bottom. The final results of protein folding for some proteins seem to be influenced more by kinetic factors than by thermodynamic ones.
FIG 1: The protein folding funnel, which illustrates how the shape of a protein affects its overall free energy.
The diagram is color coded based on the polypeptide chain's configurational entropy level (red for high entropy and blue for low entropy). From a disorderly state, folding progresses to steadily more organized conformations with reduced energy levels. Local energy minima serve as a representation for potential folding intermediates. It is believed that the native state, located near the base of the funnel, corresponds to a certain shape (or a very homogeneous set of conformations in fast exchange).
The reversible interaction of many subunits to generate an oligomer protein, which is then dependent on protein concentration, is predicted to obey the law of mass action. This is a sequence that results in the energetic and structural averaging of all the molecules in the ensemble through the fast interchange of subunits between oligomers [5]. However, major violations from the law of mass action have been found in the reversible subunit dissociation of several oligomers under hydrostatic pressure [6]. This odd behavior ranges from a reduced need for protein concentration for component association and dissociation in trimers and tetramers to a complete absence of need on protein concentration for the interaction of large protein aggregates, like virus particles [7]. The behavior of the dimer Triose phosphate Isomerase (TIM) has been thoroughly studied. It was discovered that, in apparent contradiction of the law of mass action, the dissociation of TIM subunits caused by hydrostatic pressure was entirely independent of protein concentration. The kinetics and equilibrium of the dissociation/unfolding of TIM caused by guanidine hydrochloride were examined in further experiments [8]. These experiments demonstrated that the activation free energy barriers for TIM dimer dissociation are quite high (v99 kJ/mol), which corresponds to a typical dissociation period of 15 hours or longer. The typical dissociation time is the rate limiting phase in a cycle of dissociation and association since the refolding/association of TIM subunits was discovered to be rapid.
Protein molecule individuality and conformational diseases: Transmissible spongiform encephalopathies, Alzheimer’s and Parkinson’s diseases and AD are among the serious human diseases referred to be conformational disorders. The conformational transformation of specific proteins or protein fragments from non-toxic to toxic forms is thought to be related to these illness conditions.
Conclusion
Patients with transmissible spongiform encephalopathies, such as Creutzfeldt-Jakob Disease (CJD) in humans and Bovine Spongiform Encephalopathy (BSE) in animals, develop an abnormal protease resistant form of the prion protein that builds up in the brain. Given that nucleic acids are not involved in the hereditary transmission of mammalian prion disorders, PrP appears to be the only factor in their transmission.
References
- Smith CA. How do proteins fold? Biochem Educ. 2000;28(2):76-79.
[Crossref] [Google Scholar] [PubMed]
- Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65:44-45.
- Baker D, Agard DA. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33(24):7505-7509.
[Crossref] [Google Scholar] [PubMed]
- Subramaniam V, Steel DG, Gafni A. In vitro renaturation of bovine β-lactoglobulin a leads to a biologically active but incompletely refolded state. Protein Sci. 1996;5(10):2089-2094.
[Crossref] [Google Scholar] [PubMed]
- Subramaniam V, Bergenhem NC, Gafni A, et al. Phosphorescence reveals a continued slow annealing of the protein core following reactivation of Escherichia coli alkaline phosphatase. Biochemistry. 1995;34(4):1133-1136.
[Crossref] [Google Scholar] [PubMed]
- Rietveld AW, Ferreira ST. Kinetics and energetics of subunit dissociation/unfolding of TIM: The importance of oligomerization for conformational persistence and chemical stability of proteins. Biochemistry. 1998;37(3):933-937.
[Crossref] [Google Scholar] [PubMed]
- Erijman L, Weber G. Oligomeric protein associations: Transition from stochastic to deterministic equilibrium. Biochemistry. 1991;30(6):1595-1599.
[Crossref] [Google Scholar] [PubMed]
- Kurplus M, McCammon JA. Dynamics of proteins: Elements and function. Annu Rev Biochem. 1983;52(1):263-300.
[Crossref] [Google Scholar] [PubMed]