Original Article
, Volume: 17( 2)Polymer Scavenging and Profile of E. coli 0111:B4 Lipopolysaccharide by MALDI-TOF
- *Correspondence:
- Maria Fernanda de Mello Costa CHASP, Waikato Institute of Technology, 3240, Hamilton, Waikato, New Zealand
Tel: +64-07-834-8800; E-mail: equipecentaurovet@hotmail.com
Received Date: October 13, 2017 Accepted Date: November 7, 2017 Published Date: November 16, 2017
Citation: Maria Fernanda de MC, Ivanow A, Holtz N. Polymer Scavenging and Profile of E. coli 0111:B4 Lipopolysaccharide by MALDI-TOF. Anal Chem Ind J. 2017;17(2):123
Abstract
Lipopolysaccharide is a component of the cell membrane of Gram negative bacteria and is a pro-inflammatory substance; small amounts present in the blood can be fatal to mammals. Scavenging of lipopolysaccharide from plasma has been attempted previously but the use of molecularly imprinted polymers for this purpose has not been described before. This preliminary study utilized a single lipopolysaccharide type to investigate scavenging by molecular polymers and used Matrix Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) to detect such scavenging. Commercially available LPS from E. coli O111:B4 was subjected to MALDI-TOF analysis to detect LPS profiles in solutions containing pure LPS or solutions that had been exposed to imprinted and non-imprinted polymers with scavenging potential. The current research suggests potential for molecularly imprinted polymers to be used as scavengers of LPS, although the imprinting protocols need optimization. MALDI-TOF is a suitable method for detection of LPS profiles and LPS elimination from samples.
Keywords
E. coli; Lipopolysaccharide; Technique; Molecularly imprinted polymers; Scavenging
Introduction
Endotoxin, also known as lipopolysaccharide or LPS, is a major component of the cell membrane of Gram negative bacteria. LPS is released by bacterial cell death or proliferation [1]. Gram negative bacteria are commonly found in the gastro-intestinal tract of mammals and can release LPS into the blood stream if intestinal permeability is altered.
Once LPS gains access to the blood stream, it activates immune and inflammatory responses [2]. Doses of endotoxin as small as 1 μg/Kg of body weight are sufficient to induce shock in human adult [3]. Other mammals are also very susceptible to LPS exposure and endotoxaemia causing death.
Chronic LPS exposure has been implicated with the development of metabolic syndrome and insulin resistance leading to diabetes in humans [4] and to the development of metabolic syndrome and chronic laminitis in horses [5].
A secondary, but very important issue regarding endotoxins is the frequent contamination of pharmacological preparations. Contamination of such products is of concern because of the implications of inadvertent consumption of LPS by patients using such products. Contamination of other biological products such as those originating from plasmids (recombinant DNA) from Gram negative bacteria can interfere with research experiments and can be toxic if utilized in vivo [6].
LPS is composed of three portions: Polysaccharide O, a core oligosaccharide and lipid A. The polysaccharide allows specific antibodies to be formed against specific strains of LPS, but it is the Lipid A portion which causes activation of the inflammatory cascade [7,8].
Several investigations have been conducted trying to identify therapeutic agents that would scavenge the endotoxin from circulation, or mitigate the inflammatory effects of LPS and although several candidate substances have been tested, including some with acceptable success, no synthetic scavenger designed specifically to extract the endotoxin from circulation has been developed.
MALDI-TOF has been described as a viable method for analysis of complex lipids [9] and LPS from species other than E. coli have been studied using this method [10]. Antigen H [11] and Lipid A from E. coli have been identified using MALDI-TOF [12].
LPS detection poses a complex problem; especially since LPS has the ability to adhere to most surfaces during storage and analysis and due to ease of contamination.
Quantification by HPLC has proven inconsistent and spectrophotometry using colorimetric assays can pose the challenge of degradation of the LPS during analysis, as well as loss of analyte due to surface adhesion or increase in analytic concentration due to contamination. Although not an effective quantitative method, MALDI-TOF provides the advantage of spot drying of samples, converting the disadvantage of LPS adherence to surfaces into a desirable part of the protocol.
Research has been conducted investigating the effect of various matrices on the detection of LPS with MALDI-TOF [13,14], but no publications were found at the time of writing this manuscript regarding the use of MALDI-TOF as a method of detection of LPS after reaction with potential scavenging molecules.
Molecularly imprinted polymers (MIPs) are known for their ability to mimic antibodies, remove substances from a variety of solutions and for their stability in a range of media; however existing synthesis methods for MIPs have so far made it difficult to imprint large organic molecules. One of the objectives of this research was to investigate the ability of two polymers to scavenge LPS in vitro, using ethylene glycol dimethacrylate (EDMA) and bilirubin in the manufacturing process. The other objective of this research was to investigate the use of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) as a repeatable and reliable method for detection of LPS and its scavenging from solutions by added chemicals.
Materials and Methods
Samples
Initially the identification of the chemical profile of LPS needed to be standardized so the reaction between the polymers and LPS could be monitored and measured.
Commercially available LPS from E. coli O111:B4 (Sigma-Aldrich, product L2630) was purchased and reconstituted for validation of detection methods and for testing the scavenging ability of the two polymers.
LPS profile
The protocol for this experiment is described below (MALDI-TOF). LPS samples were diluted in deionized water at a concentration of 1% (10000 ppm) for this part of the research. Once a consistent LPS profile was obtained, LPS samples were prepared and analyzed as described below.
Trial 1
Involved the product of the reaction between a first generation MIP (MIP1); a non-imprinted polymer (NIP1) and LPS at 5 different concentrations. Profiles obtained for LPS+MIP1 and LPS+NIP1 were compared to the pure LPS profile (control).
The following dilutions of LPS were utilised: 1000 ppm; 500 ppm; 100 ppm (diluted in 100% methanol); 75 ppm and 50 ppm (diluted in methanol and water; 3:1). The following was added to 800 μl of each of the dilutions of LPS: 100 μl of imprinted polymer suspension (S1 to S5) or 100 μl of non-imprinted polymer suspension (S6 to S10) or 100 μl of deionised water (S7 to S15). The samples were stored at -4°C until analysis.
Trial 2
Involved testing time aliquots of the reaction between a second-generation MIP (MIP2); a non-imprinted polymer (NIP1) and LPS at 2 different concentrations.
The time trial was executed using the 100 ppm and 75 ppm LPS dilutions, prepared with the solvents described above. Samples were labelled alphabetically: A, B and C contained LPS 100 ppm, while D, E and F contained LPS 75 ppm.
Three, 4500 μl aliquots of each of the two LPS dilutions were placed in Griffin beakers and onto six magnetic stirring plates with stirring beads for constant agitation. 500 μl of the second generation MIP was added to aliquots A and C; 500 μl of non-imprinted polymer was added to aliquots B and D; and 500 μl of deionised water was added to aliquots E and F. The final reaction volume for each sample was 5000 μl.
200 μl aliquots were taken from the reaction beakers at times 0, 1 min, 2 min, 3 min, 4 min, 5 min, 10 min, 30 min and 60 min and placed in Eppendorf tubes. The time aliquots were snap-frozen and stored at -4°C until analysis.
MALDI-TOF analysis
All samples were analysed in triplicate and both the operator and the scientists conducting the analysis of the results were blinded as to what the samples contained.
Experimental LPS samples from each of the trials were spotted onto a 100-well MALDI-TOF plate immediately after being vortex-mixed for 1 min. 1 μl of sample was spotted on each well and allowed to air dry. Once the sample was dry, 1 μl of matrix (6-aza-2-thiothymine-ATT-dissolved 10 mg/ml in acetonitrile/water 1:1 v/v) [13-15] was spotted directly on top of the dried sample and also allowed to air dry. Control and blank samples were also spotted in the same fashion.
Once samples were dry, the plate was inserted into a Voyager-DE MALDI-TOF mass spectrometer (Applied Biosystems Voyager-DE Pro). Samples were then subjected to 100 laser shots. MALDI settings were: Laser intensity: 2674; Negative ion mode and linear; 20,000 V. Mass range was set to 1000 Da to 4000 Da, which gave the best LPS profile. Samples showed good resolution and the mass profiles matched current literature for LPS MALDI-TOF profiles.
Molecularly imprinted polymers (MIPs)
A complex between LPS and bilirubin as the functional monomer was generated and replication of the complex in a polymer format was done using conventional cross-linkers (ethylene glycol dimethacrylate [EDMA]). The EDMA matrix produces a monolithic powder after initial synthesis and processing has been completed. The formation creates a lock and key model characteristic in which the polymer is formed in the presence of the target species (LPS) as a template. Once formed the target species is removed. This creates a series of binding sites which are characterised and ‘shaped’ to bind selectively to LPS and ignore other compounds/molecules present.
The formulation of the MIP recipe is performed using a confidential method which reduces recipe and polymer development significantly, with recipe development being performed twice in a 24 h period. The development process resulted in the determination of a base recipe, described in Table 1.
| Reagent | Quantity (units) |
|---|---|
| LPS | 8.1 (mg) |
| Bilirubin | 11.8 (mg) |
| EDMA ABCHC 3:1 methanol:water |
0.377 (mL) 100 (mg) 5 (mL) |
* EDMA=Ethylene Glycol Dimethacrylate;
ABCHC=Azo-bis (cyclohexanecarbonitrile)
Table 1. Base recipe for LPS MIP.
LPS was treated as the limiting factor in the recipe to allow for scaling (increasing or decreasing) based on a proportional relationship between other reagents and LPS weight.
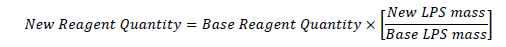
Using the proportional relationship towards the base recipe, the polymer was generated as required for testing: the reagents are mixed, combined and reacted together over a two day period, then processed for use in experimental procedures.
Results and Discussion
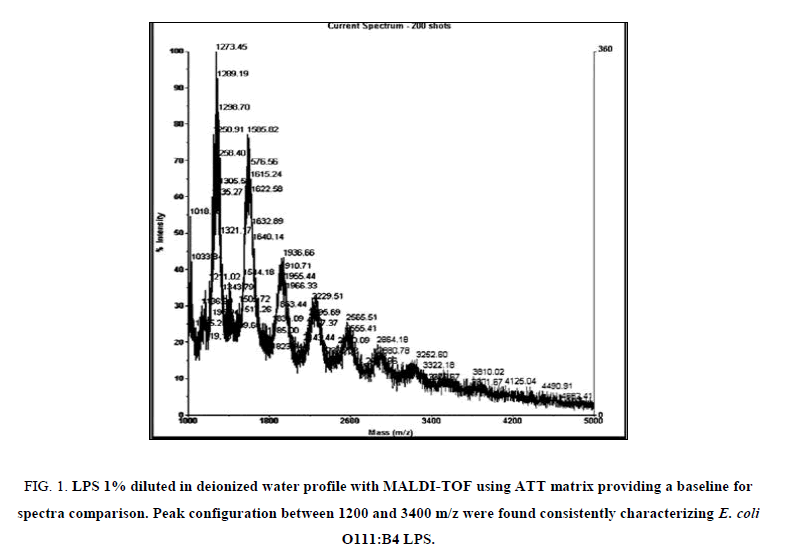
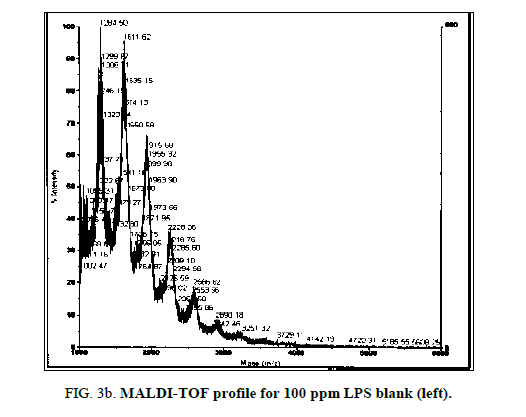
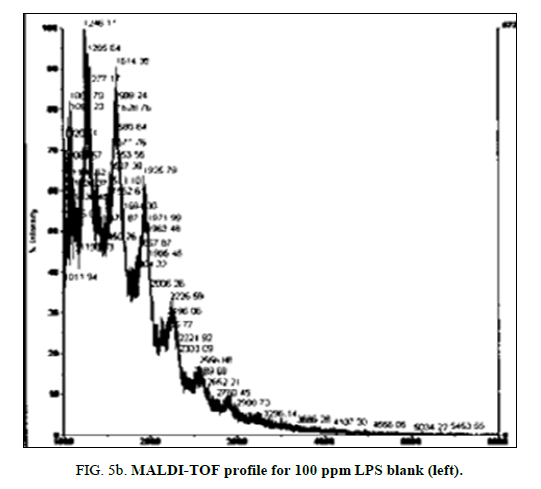
MALDI-TOF spectra for LPS with the method described above yielded a clear and consistent LPS profile (Figure 1). A cluster of mass peaks was observed between 1200 and 3400 m/z, with 4 distinct peaks at regular intervals around 1200 m/z, 1500 m/z, 1900 m/z and 2300 m/z.
Figure 1: LPS 1% diluted in deionized water profile with MALDI-TOF using ATT matrix providing a baseline for spectra comparison. Peak configuration between 1200 and 3400 m/z were found consistently characterizing E. coli O111:B4 LPS.
Trial 1
When the 1000 ppm and 500 ppm samples were analysed, spectra with clear resemblance to the LPS profile were observed, regardless of solvent used and with MIP, NIP and blank.
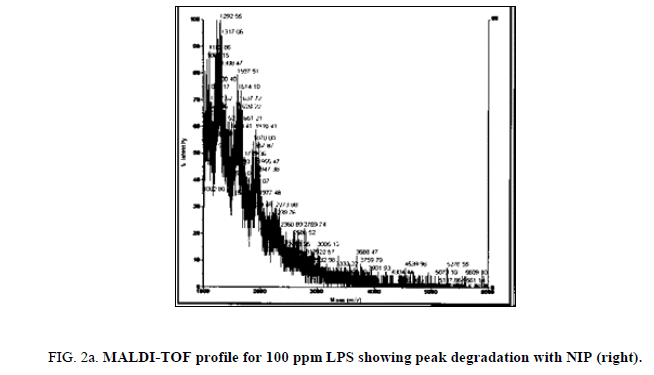
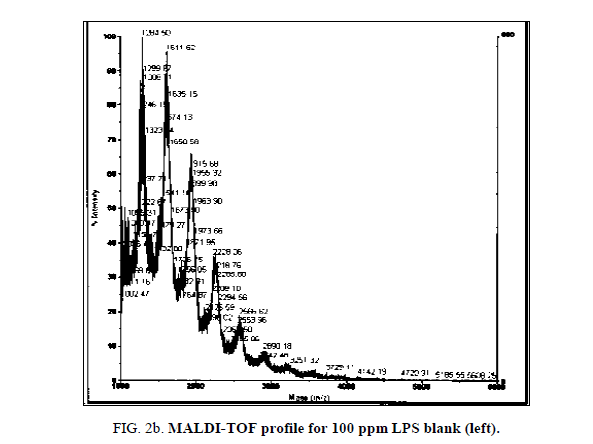
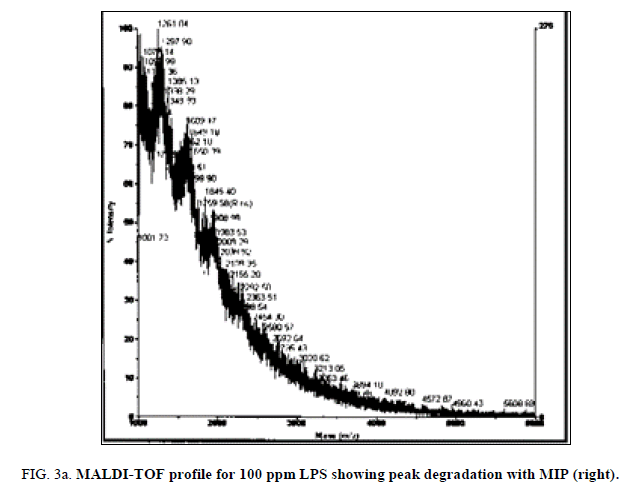
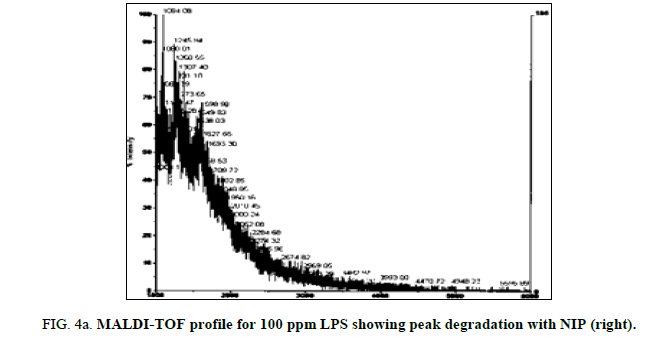
LPS peak degradation at a mass of approximately 1936 was observed both with NIP and MIP, but not with the blank at the 100 ppm dilution (Figure 2a, Figure 2b and Figure 3a, Figure 3b).
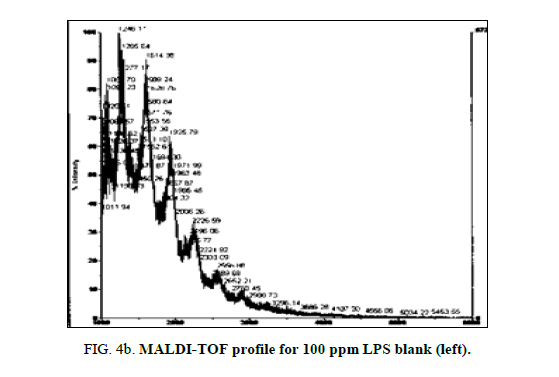
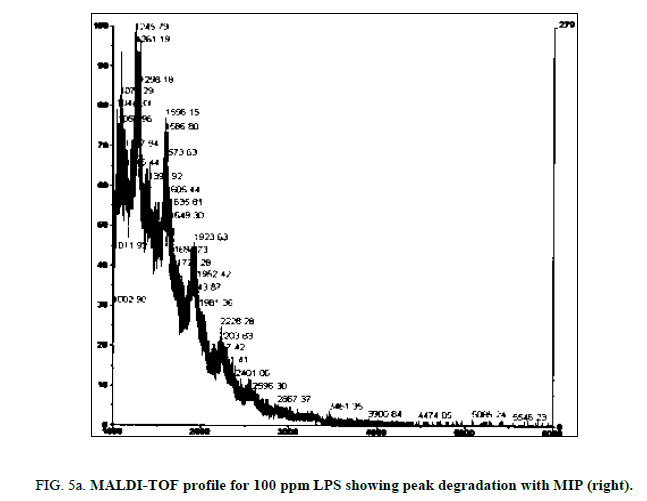
LPS peak loss was observed on the 75 ppm LPS with NIP only. Peak loss was seen at the following masses: 1925, 2226 and 2556.75 ppm LPS with MIP maintained these peaks, as did the blank (Figure 4a, Figure 4b and Figure 5a and Figure 5b).
Trial 2
During the time trial, peak loss was observed from minute 3 of the reaction between LPS and NIP (both 100 ppm and 75 ppm LPS solutions) until minute 60 (end of the time trial). The loss of peaks was more evident on the 75 ppm LPS preparation.
Peak degradation but not loss was observed in the LPS+MIP from 3 min of the time trial at the 75 ppm LPS solution only. Neither peak degradation nor loss was observed in the 100 ppm. Blank solutions displayed intact LPS profiles during the entire time trial.
In order to investigate the LPS scavenging abilities of one imprinted and one non-imprinted polymer, several methods were employed, including HPLC, spectrophotometry and MALDI-TOF. Only the MALDI-TOF protocol provided consistent and repeatable results and the data above indicates that MALDI-TOF is a reliable and consistent method for detecting and analysing LPS profiles, being a valuable method when investigating the potential scavenging of LPS by polymers and other chemicals.
Research on structural composition of strains of E. coli K12 have shown that LPS is heterogeneous but has one major and two minor glycoforms with masses between 2980 m/z and 3092 m/z [16]. In the current research purified LPS from E. coli serotype 0111:B4 was used and results show peaks with masses between 1200 m/z and 3000 m/z. The most abundant peak was detected around 1270 m/z to 1290 m/z, suggesting a monophosphorylated species.
The protocol used in this study utilised 6-aza-2-thiothymine (ATT) as the matrix, since this produced the clearer spectra, although other matrices were tested (results not shown). This is in accordance to previous publications analysing E. coli serotype 0116 which demonstrated that MALDI-TOF spectra for LPS is highly dependent on the matrix utilised [13].
In the present research, two polymers were used as potential scavengers of LPS and to test the ability of the MALDI to detect such scavenging. The malleability and rapid nature of the polymer recipe development allows for it to be a consideration for future experimentation without severe implications on the time scale, should it be required.
The initial recipe development was performed using HPLC grade pure methanol (Sigma Aldrich) due to its ability to solubilise the bilirubin monomer. The LPS complex however experiences a reluctance to solubilise in a pure methanol medium, requiring extensive mechanical action. Initial testing to develop a recipe using methanol was able to be performed, but the solubility of LPS required consideration. During the formation of the polymer both bilirubin and the target species must be able to solubilise in the chosen solvent. The extensive action required to solubilise the LPS made a pure methanol medium unsuitable and unwieldy.
To circumvent this issue, a mixture of two appropriate solvents was used. LPS dissolved easily in deionised water. Bilirubin is insoluble in water, however a diluted methanol solution in favour of methanol would still be adequate providing the water content was sufficient to solubilise the LPS. The appropriate methanol/water ratio was determined by dissolving LPS in 1 mL of water, bilirubin was added, 1 mL aliquots of methanol were introduced until both bilirubin and LPS were solubilised and remained in solution. This determined a 3:1 methanol:water solution to be the sufficient solvent.
The initial reaction creates a semi-solid polymer slurry, this is spread onto a particle sieve to facilitate drying and prepare it for grading. Grading is a two-step process which first reduces polymer ‘clumps’ and particles to an adequate size. The ground particles are then made into a slurry with excess solvent, the particles of adequate size will settle out of solution with the smaller particles being removed with the supernatant (this is then repeated to ensure efficient removal). The remaining slurry is tilled and dried. The MIP is placed into a soxhlet apparatus for 20 h removing adhered solvents and any trace quantities of unreacted reagents. The dry powder is then placed in suspension with 1 mL of 3:1 methanol:water for every 100 mg of polymer. The suspension is left to gyrate and mix overnight, allowing for the polymer to swell and pores t o open up making them more accessible to the LPS complex. This suspension was then used in the experimentation described in this report. The process from initial reaction to suspension that is ‘ready for use’ can be performed in one to two weeks.
The mixtures of polymer suspension and test solutions have been the primary applications in this project. With further research other possible mediums could be used and developed, these could include the affixation of the polymer to a filter cartridge/membrane or the manufacture of column/s containing the polymer; Though column manufacture is not recommended until experimentation has proceeded further.
The experimentation performed in this project renders the polymer ‘expendable’, meaning that it has been treated as a ‘single use’ component. In future experimentation, retention and regeneration of the polymer could be used to minimise reagent expense. This will still consume time, but should reduce overall time by limiting actual polymer generation cycles. Whether this is an appropriate course of action for the final application of the polymer is yet to be determined.
After successful imprinting in an EDMA matrix, polymers based on pyrrole will be attempted as these will be fully biocompatible. Pyrrole based polymers also have a significantly smaller effect on the time scale of polymer production and subsequent experimentation. A recipe can be determined and the polymer reacted and processed within 48 h, drastically diminishing waiting time between generation and experimentation. It can be used in the same manner of testing used during this project. It also has a range of delivery options, which could offer further and more diverse methods of testing. The formation of the pyrrole polymer does require a larger mass of the target species for minimum production however. A minimum quantity of 50 mg would be required for each batch of polymer, but the polymer could also be retrieved and regenerate between experimentation cycles. The polymer generated would also achieve a greater mass and volume than the bilirubin polymers. Even if treated as a ‘single use’ product, the total amount generated could be spread across a significantly larger quantity of tests.
In the discussion below, "peak degradation" refers to the diminishing presence of a peak or set of peaks on the mass spectra, indicating that this particular portion of the LPS is being affected during the reaction. "Peak loss" refers to the clear absence of a repeating peak on the mass spectra, indicating that this portion of LPS is no longer present in the reaction, due to being scavenged out by the polymer.
When LPS scavenging was investigated at concentrations of 1000 ppm and 500 ppm, a volume of 100 μl of polymer (both imprinted and non-imprinted) was unable to change the LPS profile as seen in the blanks. This was likely due to the fact that the polymer was either insufficient or entirely consumed before any detectable peak deterioration was observable.
MALDI-TOF proved to be a reliable and repeatable method in the detection of LPS and also documenting the effect of the reaction between LPS and different polymers. This methodology can be used in future trials researching the ability of different constructs/chemicals to bind and scavenge LPS from a solution.
Both the imprinted and the non-imprinted polymers showed the ability to deteriorate the LPS peak profile of the 100 ppm LPS solution, indicating that a reaction was occurring. Although deterioration was seen, this reaction was not sufficient to eliminate any peaks in the LPS profile, which demonstrates inappropriate binding.
Of the two polymers used, only the non-imprinted showed the ability to degrade LPS to the point where peaks were eliminated and a change in the profile was seen. Peak losses were not seen in the imprinted polymer, or in the blanks. This result indicates that the non-specific, non-imprinted polymer demonstrated ability to bind to portions of the LPS and altered its profile, suggesting ability to remove parts of the LPS molecule.
This finding can be used in the manufacturing process of the imprinted polymers in the direction of ameliorating the chemical construct of the imprinted polymer. This finding also provides the fundamentals of evidence that LPS scavenging by polymers is possible, although further research into the imprinting and specificity of the binding are required.
Conclusion
This preliminary in vitro study assessed the ability of MALDI-TOF analysis to detect an LPS profile for E. coli O111:B4 and to measure the ability of polymers to bind LPS. The results show potential for using this MIP for LPS scavenging for real-time elimination of LPS from pharmaceutical formulations. Improvement in the MIP design should lead to a third generation construct with enhanced scavenging abilities, although further research is required to investigate MIP scavenging ability of LPS in plasma in vitro.
Acknowledgement
Funding: This study was funded by the Waikato Institute of Technology (Wintec) Research Office through a Contestable Funding Grant issued to Dr De Mello Costa (Wintec) and Dr Miruna Petcu (Ligar).
Conflicts of Interest
The authors have no other actual or potential conflicts of interest.
No animals or human subjects were used in this research.
References
- Hodgson JC. Endotoxin and mammalian host responses during experimental disease. Journal of Comparative Pathology. 2006;135(4):157-175.
- Copeland S, Warren HS, Lowry SF, et al. Inflammation and the host response to injury investigators. Acute inflammatory response to endotoxin in mice and humans. Clinical and Diagnostic Laboratory Immunology. 2005;12(1):360-67.
- Warren HS, Fitting C, Hoff E, et al. Resilience to bacterial infection: difference between species could be due to proteins in serum. The Journal of Infectious Diseases. 2010;201(2):223-32.
- Moreno-Navarrete JM, Fernández-Real JM. Antimicrobial-sensing proteins in obesity and type 2 diabetes. Diabetes Care. 2011;34(2):335-41.
- Forbes G, Church S, Savage CJ, et al. Effects of hyperimmune equine plasma on clinical and cellular responses in a low-dose endotoxaemia model in horses. Research in Veterinary Science. 2012;92(1):40-4.
- Magalhães PO, Lopes AM, Mazzola PG, et al. Methods of endotoxin removal from biological preparations: A review. J Pharm Pharm Sci. 2007;10(3):388-404.
- Netea MG, van Deuren M, Kullberg BJ, et al. Does the shape of lipid A determine the interaction of LPS with Toll-like receptors? Trends Immunol. 2002;23(3):135-9.
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annual Review of Biochemistry. 2002;71(1):635-700.
- Schiller J, Süß R, Arnhold J, et al. Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Progress in Lipid Research. 2004;43(5):449-88.
- Pupo E, Hardy E. Complexity and solutions to the isolation problem of Gram negative lipopolysaccharides’ bacteria molecular species. Biotecnol Apl. 2009;26(1):78-84.
- Chui H, Chan M, Hernandez D, et al. Rapid, sensitive and specific E. coli H antigen typing by matrix-assisted laser desorption ionization-time of flight-based peptide mass fingerprinting. Journal of Clinical Microbiology. 2015;53(8):2480-85.
- Hankins JV, Madsen JA, Needham BD, et al. The outer membrane of Gram-negative bacteria: Lipid A isolation and characterization. Bacterial Cell Surfaces: Methods and Protocols. 2013:239-58.
- Zhou P, Altman E, Perry MB, et al. Study of matrix additives for sensitive analysis of lipid A by matrix-assisted laser desorption ionization mass spectrometry. Applied and Environmental Microbiology. 2010;76(11):3437-43.
- Stu?biger G, Belgacem O, Rehulka P, et al. Analysis of oxidized phospholipids by MALDI mass spectrometry using 6-aza-2-thiothymine together with matrix additives and disposable target surfaces. Analytical Chemistry. 2010;82(13):5502-10.
- Mares J, Kumaran S, Gobbo M, et al. Interactions of lipopolysaccharide and polymyxin studied by NMR spectroscopy. Journal of Biological Chemistry. 2009;284(17):11498-506.
- Müller-Loennies S, Lindner B, Brade H. Structural analysis of oligosaccharides from lipopolysaccharide (LPS) of Escherichia coli K12 strain W3100 reveals a link between inner and outer core LPS biosynthesis. Journal of Biological Chemistry. 2003;278(36):34090-101.