Research
, Volume: 13( 3)Populations of Biomphalaria spp. Snails Isolates in Kenya Show Variable Resistance to Schistosome Infection
i DM1, Githui EK2*, Aman RA3, Wanjala FM1, Stella RK4, Njiru PN5, and Nguu EK61Department of Biological Sciences, University of Eldoret, Eldoret, Kenya
2Molecular Genetics and Biotechnology Lab, Institute of Primate Research, Karen, Nairobi, Kenya
3African Center for Clinical Trials, Nairobi, Kenya
4Department of Biological Sciences, Maasai Mara University, Narok, Kenya
5Department of Biochemistry, University of Nairobi, Nairobi, Kenya
6Department Agricultural Resource Management, University of Embu, Embu
- *Correspondence:
- Elijah Kem Githui , Molecular Genetics and Biotechnology Lab, Institute of Primate Research, Karen, Nairobi, Kenya, E-mail: kegithui@yahoo.com
Received: September 27, 2018; Accepted: October 04, 2018; Published: October 09, 2018
Citation: Ngigi DM, Githui EK, Aman RA, et al. Populations of Biomphalaria spp. Snails Isolates in Kenya Show Variable Resistance to Schistosome Infection. Res Rev Biosci. 2018;13(3):144.
Abstract
Fresh water snails of genus Biomphlaria, Bulinus and Ocomelinia are important vectors for human schistosomiasis. Human contact patterns with water infested with cercaria are important factors in transmission of schistosomiasis and these conditions are associated with tropical climate wetlands. However, some snail strains within the genera that transmit cercaria are resistant to infection, providing means where these traits can be harnessed for biological control. PCR rDNA probes were used to determine miracidia/cercaria infected Biomphalaria snail isolates within Kenya. The derived sequences together with similar Genbank datasets were applied in phylogenetic analysis. PCR results showed that susceptibility/resistance trait occurs in proportion of the field sampled snails and also those maintained in laboratory culture. Phylogenetic analysis of sampled snails in this study together with composite dataset of similar rDNA sequences across Africa showed that Biomphalaria spp population structure is composed of distinct monophyla lineages and clusters of closely related isolates or clones. Further, the analysis indicated that nomenclature of Biomphalaria spp. classification needs revision. This provides evidence for resistance to cercaria transmission in some isolates of Biomphalaria spp. within Kenya but the trait does not cluster together in phylogenetics that is based on rDNA gene.
Keywords
Snails; Phylogeny; Schistosomiasis; Resistance
Introduction
Fresh water snails of genus Biomphlaria, Bulinus and Oncomelania are important vectors for human schistosomiasis. The disease also infects vertebrate mammals including primates, cattle, birds and crocodilians [1]. In human, the chronic disease (Bilharziasis) is debilitating in early childhood, therefore the main impact of schistosomiasis is low productivity in terms of human resources.
World Health Organization (WHO) global epidemiological data shows that schistosomiasis is endemic in 78 tropical and subtropical countries with 779 million people are at risk of schistosomiasis. More than 230 million people are infected with 20 million suffering from debilitating illnesses [2,3]. Due to the association of the disease incidence with poverty, Africa accounts for majority of the infections.
The life cycle of schistosomes has initial developmental stages in the invertebrate snails and further developments and maturity in a vertebrate host. Schistosome eggs that are released into fresh water by vertebrate host hatch to miracidia larvae that infect snails. After several weeks of sporocysts asexual multiplication in the snail tissue, the next larva called cercaria, are released into the water where they infects vertebrate host upon contact. Therefore, snail infection and human contact patterns with water infested with cercaria are important factors in transmission of schistosomiasis [4,5]. Therefore, ecological conditions favoring transmission cycle are tropical climate, fresh water wetlands, presence of intermediate snail and lack of proper sanitation. Wetlands provide a valuable focal point for domestic water consumption, irrigation agriculture and fishing dams, as well as source of water for wild and domestic animals [6]. Farming wetlands are valuable but also focal point of disease transmission [7,8].
The species of schistosomes of economic importance include Schistosoma mansoni, originally from Africa and later spread to South America; S. haematobium, found in Africa and adjacent Middle East regions and S. japonicum found in South-East Asia. S. intercalatum and S. mekongi have more localized geographic location while S. bovis is an important parasite for angulates [9,10]. Fresh water snails of the genus Oncomelania are the important vectors for S. japonicum while the pulmonate snails of the genus Bulinus and Biamphalaria transmit S. haematobium and S. mansoni, respectively [11].
Biological control using competitor fresh water snails in the reduction of schistosomiasis transmission has achieved appreciable successes in the Caribbean Islands and in Brazil [12]. In their studies in West Indies, Pointier et al. and Guimareas et al. showed that B. glabrata and B. straminea can be eliminated using competitor thiarid snail, Melanoides tuberculata [13,14]. Other approach in biological control of transmitting snails is by exploiting snails genetically resistant to schistosomes. B. glabrata and B. tenagophila are important in transmission of S. mansoni in Brazil, but the Taim strain of B. tenagophila consistently shows absolute resistance against the parasite [15]. Several studies on challenge infection of Taim strain with miracidia shows systematic resistance of this strain to S. mansoni [16,17] and the resistance trait was dominant during crossbreeding with susceptible B. tenagophila strains [18].
In the African region, it has been noted that not all species within the genus B. africanus act as intermediate host for S. haematobium and similarly, S. mansoni miracidia infects only certain strains of B. glabrata and B. ugandae species while some strains of are refractory to infection [19]. This study explores population variability of Biomphalaria snails samples from within Kenya and tests PCR probe for cercaria transmission to determine the susceptibility or resistant trait in these snails.
Methods and Materials
Sampling and specimen identification
Field work sampling of Biamphalaria snails was carried out in two schistosomiasis endemic regions: Mwea irrigation farmlands that drain from Mt Kenya in central Kenya highlands and another site, Kibwezi dam in Eastern part of Kenya towards the coastal region. Sampling involved a technologist from Institute of Primate Research (IPR) schistosomiasis program to guide in determining microhabitat sites and vector snails by morphology. Snails were collected alive and placed on wet cotton wool in perforated containers then transported to snail culture facility at IPR within 24hrs.
Snail infection with miracidia
To standardize infections, snail were put in de-clorinated tap water in 200 ml beakers containing miracidia (at 5/ml-10/ml) that had been obtained from stool from on-going schistosomiasis experiments in non-human primates at IPR. After period of 3 to 6 weeks of incubation, cercaria shedding was done by procedure of direct light illumination where individual snails were put in 10ml water in beaker and cercaria shedding determined by observation on 10 X magnification stage microscope.
DNA extraction and preservation
Biopsy tissues from snails were obtained and subjected to DNA extraction using Qiagen kit (Qiagen Inc. MD, USA) to maximize yield. Briefly, 20 μl Proteinase K and 200 μl of digestion/lysis buffer was added to sample and incubated at 56ºC water-bath for 1hr. The lysate was pelleted to remove debris by centrifugation (X1000 g) and the supernatant aspirated into silica matrix spin column to bind DNA. After initial centrifugation at 10,000 g for 2 min the DNA in the column matrix was 2X cleaned in wash buffer centrifugations then eluted in 200 μl nuclease free distilled de-ionized water (ddH20), as described in owner’s manual. DNA was visually analyzed on 1% agarose gel with Ethdium bromide under utra-violet illumination and quantity determined by 260/280 nm ratio. The DNA samples were labelled and stored at -20ºC for subsequent analysis.
Polymerase chain reaction (PCR) amplification
PCR was carried out using specific primers dependent on target gene of the extracted DNA, Schistosoma spp cercaria and Biomphalaria spp ribosomal target, respectively (TGCATACTGCTTTGAACATTC; CCTGACTAGGCTGGT) (TCGAAGCGCACGAACGCG; GGAAGGATCATTAAAGGCTT). The PCR amplification was done using approximate 70ng/ul of the template DNA, 0.5 μM of each primer, 200 μM each dNTP, 10 X buffer with 2.0 mM MgCl2 and 0.5 U Takara Taq polymerase, made up to 25 μl with ddH2O. Cycling conditions were: Taq polymerase activation 92ºC, 3 min; then 30 cycles profile: denaturation, 92ºC, 1 min; annealing 52ºC-57 ºC, 1 min; polymerase nucleotide elongation 70 ºC, 1 min; final polymerase extension step 70ºC, 3 min and final storage in the PCR machine at 8ºC.
Sequencing and phylogenetic analysis
Gene clean procedures on the PCR products were carried out according to innuPREP procedures (Lifescience Jena, Germany) and samples sent for custom sequencing (Macrogen Inc., Denmark). The raw sequences were analyzed and edited in BioEdit Suite (BioEdit v7.0.5, Tom Hall Ibis Therapeutics) to get consensus sequences and trim 3’ and 5’ends. Verification for cercaria derived sequences was infection was previously done via BLAST analysis to determine highly related sequences in NCBI nucleotide database. In another set of gene clean and sequencing procedures, Biomphalaria rDNA ITS1 sequences were deposited via BankIt procedures in NCBI to GenBank (Accession: 579768.1-579823.1). To determine snail species, subspecies and strains population structure, analysis was performed in Phylip tree [20] and test of neutrality tested using Tajima-D [21] using program implemented in MEGA Version 6 software [22].
Results
Standardized sensitivity PCR detection of cercaria
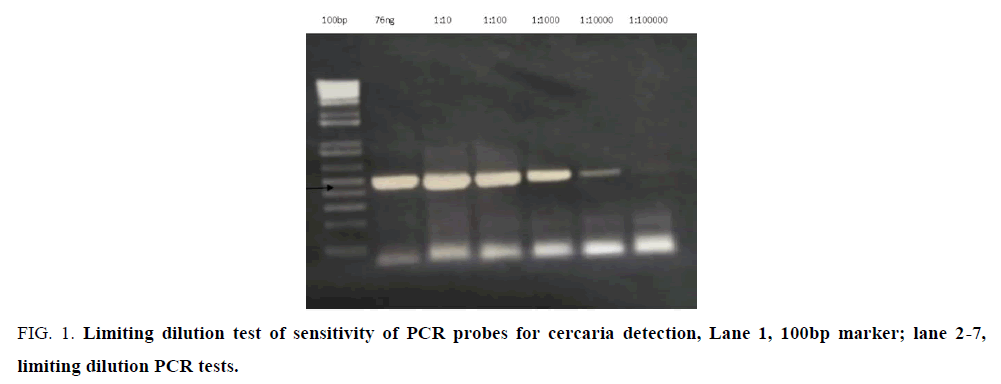
PCR done on cercaria DNA using pair of primers franking the ITS 1 schistosoma rDNA yielded a distinct band approximate 500bps (FIG.1, arrow). Detection tested by limiting dilution was sensitive at picoMole/ femtoMole concentrations (1 × 10-12-1 ×10-15g) of DNA. The PCR product had previously been verified through sequencing to be schistosome rRNA gene.
Field PCR screening of cercaria infection in snails
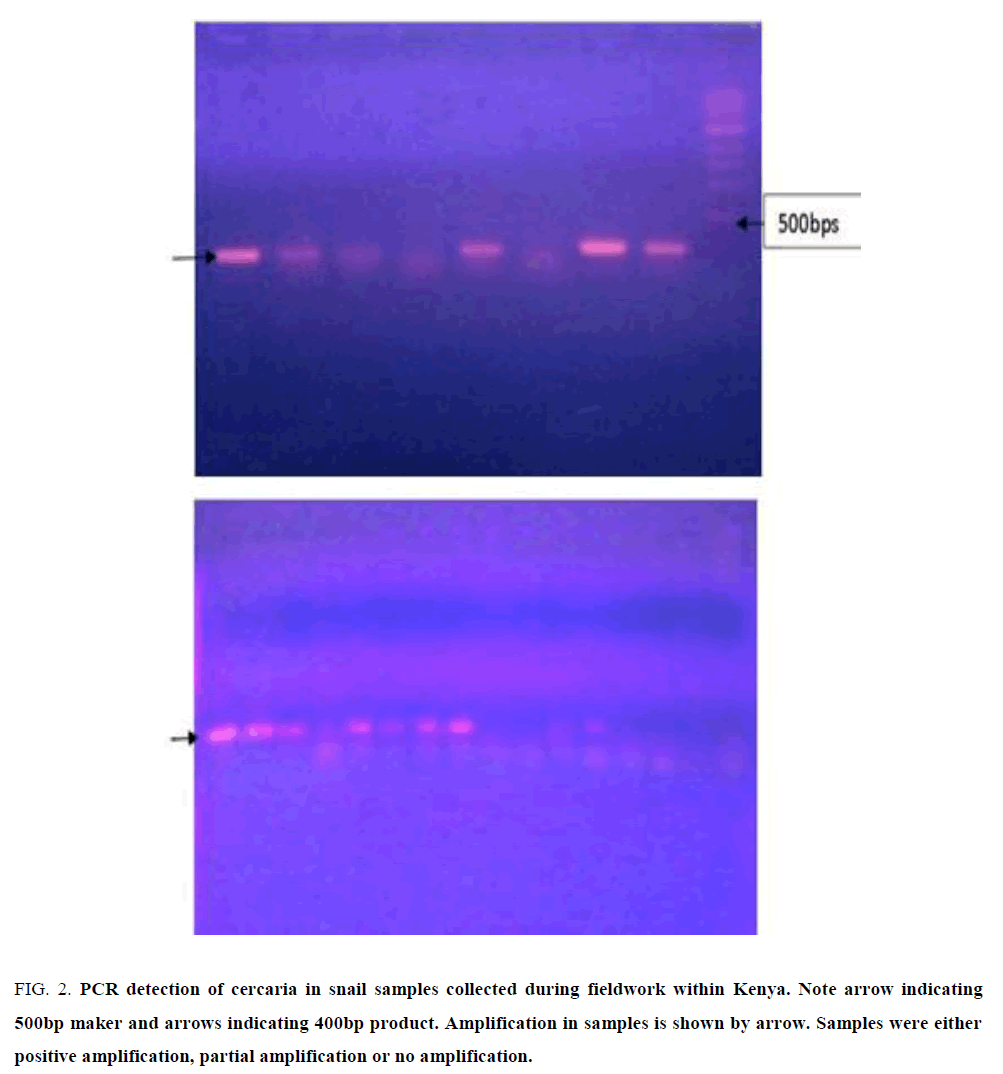
PCR on DNA extracted from snails after exposure to infection by miracidia produced specific amplification of target size band of approximately 400 bps in rDNA ITS gene (FIG. 2, arrow) but there were amorphous amplification, possibly degraded target. In the target amplification, the results were not uniform, with some snails demonstrating higher positive titre of sporocyst/cercaria DNA while others had limiting titre while others were negative.
Figure 2: PCR detection of cercaria in snail samples collected during fieldwork within Kenya. Note arrow indicating 500bp maker and arrows indicating 400bp product. Amplification in samples is shown by arrow. Samples were either positive amplification, partial amplification or no amplification.
Phylogenetic structure of Biomphalaria Snails in Kenya
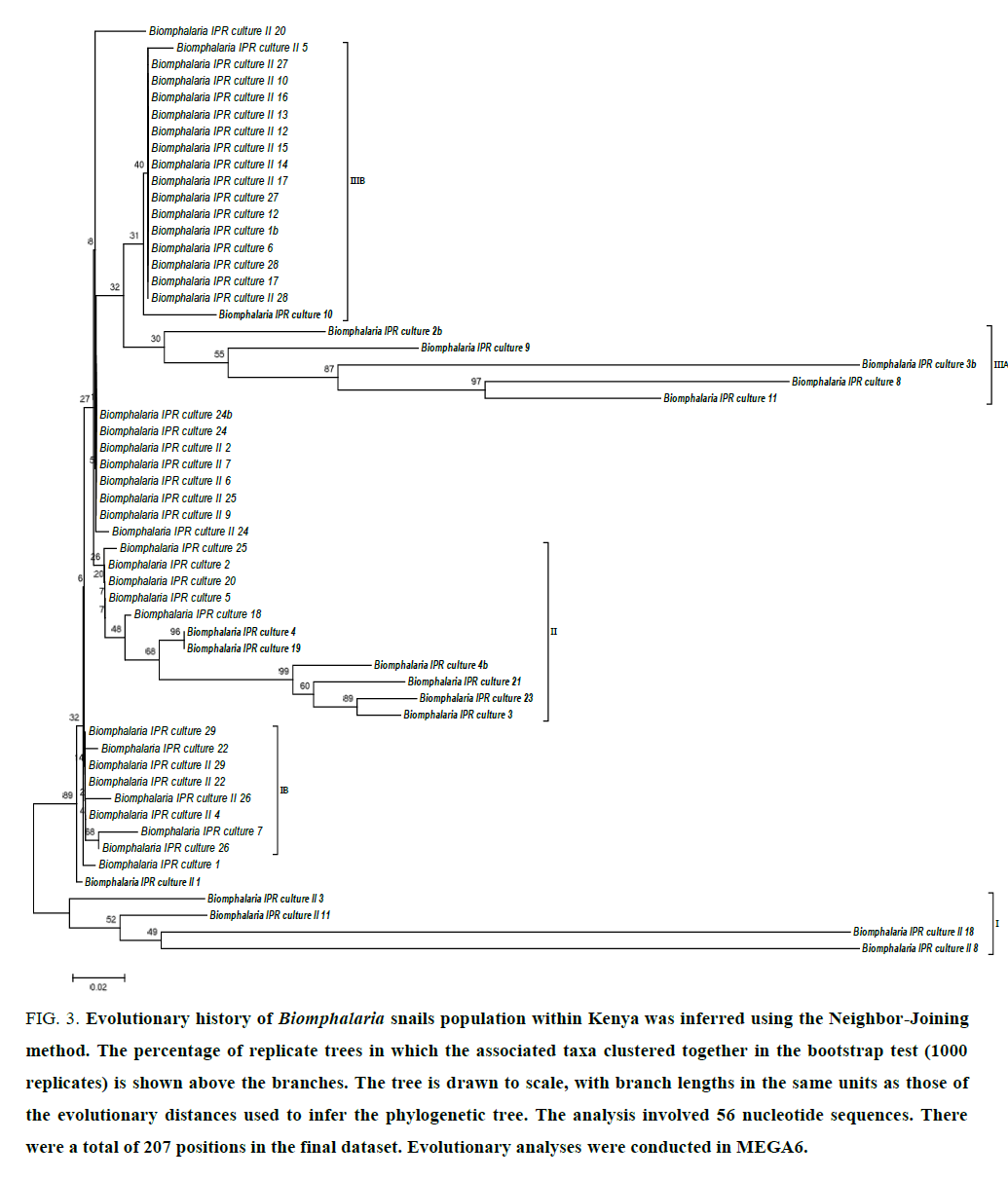
The 450 bps PCR products obtained using primers for Biomphalaria rDNA 5.8 s, ITS1 were sent for custom sequencing and edited sequences subsequently submitted to GenBank (Accession: 579768.1-579823.1). The phylogenetic structure showed that certain lineages of Biomphalaria are monophyletic with long distance time separation lineages I, II and IIIA (FIG. 3) while the majority of the sampled snails populations clusteres were phylogenetically indistinguishable (IB, IIIB and other clonal off shoots within cluster II and III).
Figure 3:Evolutionary history of Biomphalaria snails population within Kenya was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown above the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 56 nucleotide sequences. There were a total of 207 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
Phylogeographic structure of Biomphalaria snails within Africa
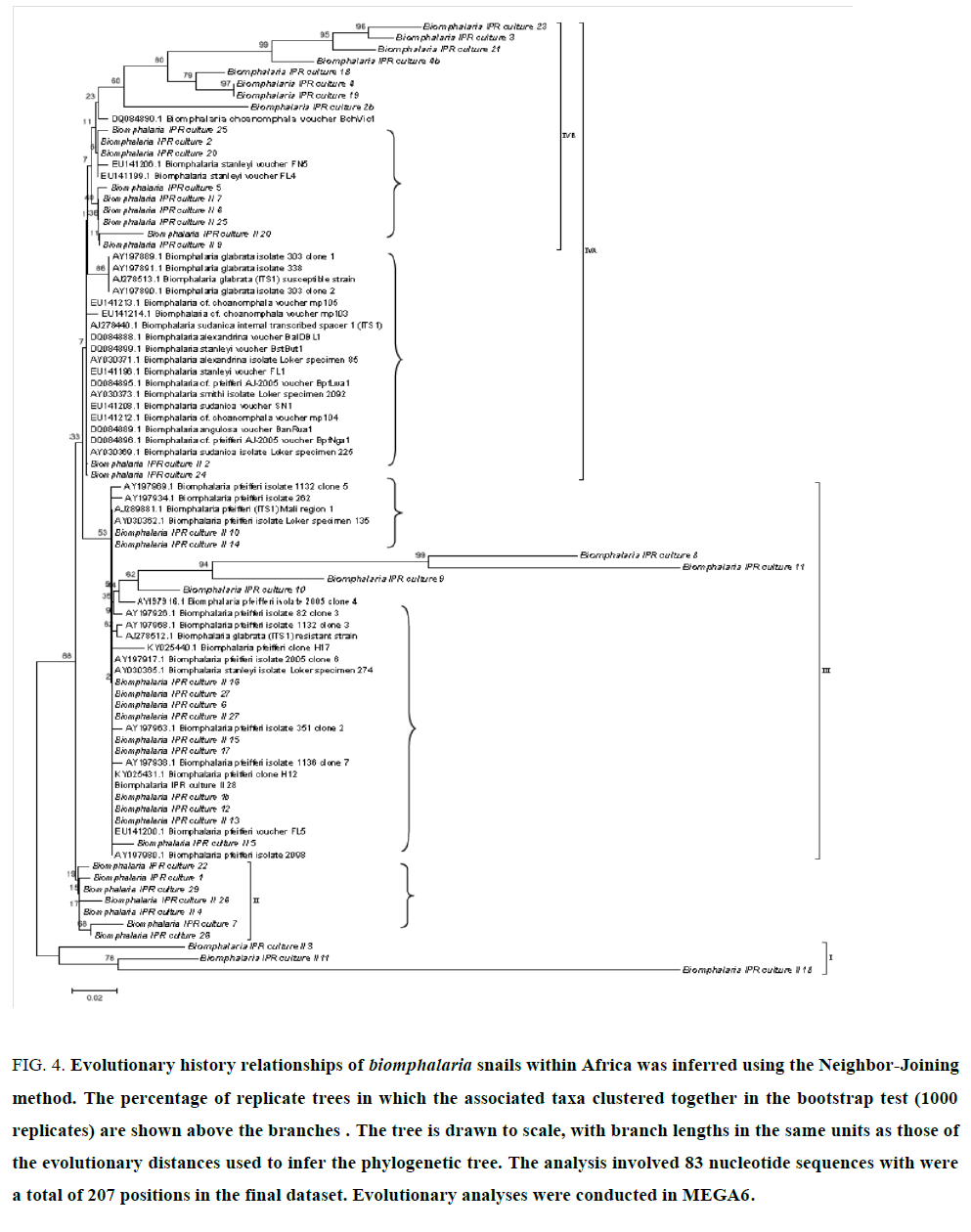
Snails rDNA sequences obtained in this study together with similar sequences datasets from across Africa obtained in GenBank showed that most snail are clusters of closely related populations, isolates or are derived from clonal propagation (FIG. 4, curly brackets) with few monophyletic lineages (bold). While nomenclature may need revision, four clusters are evident (brackets I, II, III, and IV). B. pfeifferi is the most abundant with other representative populations in B. sudanica, B. choanomphala, B. stanleyi, B. alexandria and B. glabrata. Majority of Kenyan sampled snails (italized) belong to B. pfeferri but there is ample representation in the other clusters.
Figure 4:Evolutionary history of Biomphalaria snails population within Kenya was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown above the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 56 nucleotide sequences. There were a total of 207 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
Biomphalaria snails population drift
Test of drift from neutrality was performed using Tajima’s D. When population is at equilibrium neutrality, the nucleotide diversity(π)and the number of nucleotide segregating sites (Θ) are indistinguishable. In the tested Biomphalaria populations (TABLE 1), Θ is significantly greater than π resulting in pronounced negative Tajima’s D.
| Population | m | S | ps | Θ | p | D |
|---|---|---|---|---|---|---|
| Kenya regional Biomphalaria | 56 | 146 | 0.705314 | 0.153542 | 0.080617 | -1.681654 |
| Africa composite Biomphalaria | 83 | 131 | 0.632850 | 0.126823 | 0.051698 | -2.003619 |
Note: The analysis involved 56 and 83 nucleotide sequences representation of Kenya and Africa regions, respectively. There were a total of 207 positions in each of the final dataset. m=number of sequences, n=total number of sites, S=number of segregating sites, ps=S/n, Θ=ps/a1, p=nucleotide diversity, and D is the Tajima test statistic (p and S/a1 both estimate Θ, where E (expected) E [p]=Θ, E [S]=a1 Θ), software default significant at P < 0.10. Evolutionary analyses were conducted in MEGA6.
Table 1: Tajima's Neutrality Test for Biomphalaria snails populations.
Discussion
Human population growth and erratic weather patterns have prompted the continued expansion of irrigated agricultural schemes in Africa hence transmission of schistosomiasis in this region is expected to increase [7,8]. Moreover, there is no effective vaccine against schistosomiasis [23,24]. It is therefore, important that various public health control measures for schistosomiasis are carried out in concert.
Freshwater Biomphalaria snails are the vector for Schistosoma mansoni and strategies to interrupt the disease at vector stage need more emphasis [25]. These snails show variability in susceptibility to transmit the human infective larvae stage, cercaria and some studies implicate macrophage-like hemocytes cellular immunity for the resistance to infection [26-28]. A necessary aspect in determining transmission is provision of tools to detect infection of snails by cercaria. The traditional demonstration of snail infection via shedding of cercaria upon direct illumination is easy and inexpensive but requires long and variable pre-patent periods from three weeks to two month for snails to readily shed [29]. Molecular detection of snail infection by cercaria has been done using PCR targeting NADH subunit 5 gene, Cytochrome c oxidase gene and rDNA ITS regions [30,31]. In this study we applied PCR primers franking schistosomes rDNA ITS1 that were specific and highly sensitivity (FIG.1), demonstrated at femtomole concentrations similar to detection levels obtained by [30]. Much higher sensitivity can be achieved by designing nested primers for the target DNA fragment and this can provide a simple schistosomes infection detection PCR kit for epidemiological survey of schistosomiais transmission [32].
Isolates of Biomphalaria spp and Bulinus spp snails vary in their ability to transmit schistosomes based on their susceptibility or resistance traits that is sometimes ascribed to hemocyte immune cells [26,33,34]. For example, S. mansoni miracidia infects only some strains of B. glabrata and B. ugandae while some other strains are refractory to infection. Similarly, it has been noted that not all species within the genus Bulinus africanus act as intermediate host for S. haematobium [19,35]. Snail culture at our IPR laboratory used in this study composed of different species and strains sampled from across Kenya. We applied PCR primers targeting schistosomes rDNA ITS region to detected the pre-patent infections of the snails by cercaria. The results (FIG. 2) show that not all the snail samples that had been exposed to miracidia sustained infections and the titres of the pathogen were different. Some snails were miracia/cercaria free and the blurred smear could be poor amplifications in the degraded pathogen. This indicates that some variant snails in every sampling resist schistosome infections. In fact, after long term culture of snails exposed to miracidia infection, the resistant snails become the dominant population.
Snails that transmit human schistosomiasis in Kenya are Bulinus spp, for S. haematobium that is mainly found in the coastal region and the adjoining eastern regions and Biompalaria spp, for S. mansoni in the central and western parts of the country although there are intermixtures of both species in some regions [19,35,36]. Detailed studies on phylogenetics and classification systematics for these snail vectors of schistosomiasis is an important prerequisite in defining isolates and strains that are susceptible or resistant to schistosomes [30,37]. Species and strains identification using the snail shell are not completely reliable hence the need for revised nomenclature combining morphological features and molecular markers [19,36].
In this study, phylogenetics analysis of Biomphalaria isolates using rDNA ITS sequences identified at six clusters (FIG. 3) within Kenya with some lineages that are monophyletic over long evolutionary time (cluster I, II and IIIA) while other populations reflected closely related clonal progenies (cluster IB, IIB) that are offshoots from these monophylectic lineages. This characteristics is similarly observed in the composite phylogenetic analysis of Biomphalaria spp from across Africa drainages (FIG. 4) suggesting that some field isolates are likely to be monophyla lineages (in bold) while other clades are clonal or very closely related isolates (FIG. 4, curly brackets). The nomenclature while representative of the various isolates of Biomphalaria spp appears uncoordinated and may need revision [19]. Kenya isolates used in this study (italized in FIG. 3,4) show diverse genetic structure representative of six major isolates from across Africa comprising of B. pfeiferri, B. sudanica, B. choanomphala, B. stanleyi, B. alexandria and B. glabrata (FIG. 4, brackets I, II, III, IVA and IVB). This is possibly due to Kenya’s geographic location in relation to the main drainages in Africa: Lake Victoria and the Nile basin, river Congo basin and drainages from Eastern and Southern Africa Highlands to Indian Ocean. The disruption and redistribution of river systems with the formation of East African Rift Valley in Miocene period provides a guide to tropical Africa fauna phylogeographic affinities in this region [38,39].
Population structure of these fresh water snails is likely the result of isolated wetlands separated by geographic barriers. Tajima neutrality test gave significant negative values (-1.681654 and -2.003619) for Kenyan and Africa regional isolates, respectively (TABLE 1). Tajima-D values close to zero indicate that the nucleotide diversity is near neutrality and the population from which samples were drawn is almost in equilibrium with respect to drift and mutation [21]. The datasets showed negative Tajima-D value implying purifying selection against certain deleterious alleles or population expansion where new selected-for alleles are still in low frequency resulting in low heterozygozity. The results indicate that the isolated wetland habitat for freshwater snails possibly represents snail populations undergoing recent expansions.
Conclusion
Biompalaria snail populations sampled in this study comprise of monophyletic lineages and clusters of closely related isolates or clonal expansion. Similar population structure was observed when Africa regional sequence datasets were analyzed.
The PCR detection of pre-patent cercaria infection showed that some snails are susceptible while others were resistant to schistosomes infection.
Acknowledgement
This study was funded by grant by National Council for Science Innovation and Technology (Nacosti, Kenya) to Study Schistosomiasis Transmission in Kenyan Wetlands (2013-2016) where EKG was the Principle Investigator. This results analyzed in this manuscript will contribute to the ongoing Ph.D. dissertation for David M Ngigi.
References
- Bowles J, Blair D, McManus DP. A molecular phylogeny of the human schistosomes. Mol Phylogenet Evol. 1995;4:103-9.
- WHO (2014). Weekly epidemiological record Releve epidemiologique hebdomadaire. Janvier. 2014;89:21-8.
- Chitsulo L, Engels D, Montresor A. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41-51.
- Neva FA, Brown HW. Blood flukes of human biengs. In basic clinical parasitology, Norwalk, Connecticut: Appleton and Lange 1994;245-62.
- Hamburger J, Xu YX, Ramzy RM, et al.Development and laboratory evaluation of a polymerase chain reaction for monitoring Schistosoma mansoni infestation of water. Am J Trop Med Hyg. 1998;59:468-73.
- Bigard C, Pioch S. The inclusion of biodiversity in environmental impact assessment: Policy-related progress limited by gaps and semantic confusion. J Environ Manage. 2017;200:35-45.
- Wilson RA, Coulson PS. Antibodies to glycans dominate the host response to schistosome larvae and eggs: is their role protective or subversive? Mem Inst Oswaldo Cruz. 2006;101:13-20.
- Wilson RA, Coulson PS. Immune effector mechanisms against schistosomiasis: Looking for a chink in the parasite's armour. Trends Parasitol. 2009;25:423-31.
- Barber KE, Mkoji GM, Loker ES. PCR_RFLP analysis of the ITS2 region to identify schistosoma haematobium and S. bovis from Kenya. Am J Trop Med Hyg. 2000;62:434-440.
- Despres L, Kruger FJ, Imbert-Establet D, et al. ITS2 Ribosomal RNA indicates Schistosoma hippopotamia is a distinct species. Int J Parasitol. 1995;25:1509-14.
- Sturrock RF. Schistosomiasis epidemiology and control: How did we get here and where should we go? Mem Inst Oswaldo Cruz. 2001;96:17-27.
- Pointier JP, Giboda M. The case for biological control of snail intermediate hosts of Schistosoma mansoni. Parasitol Today. 1999;15:395-397.
- Pointer JP, Guvard A, Mosser A. Biological control of Biomphaalaria glabrata and B straminea by the competitor snail Thiara tuberculata in a transmission of schistosomiasis in Martinique, French West Indies. Ann Trop med Parasitol. 1989;83:263-69.
- Guimaraes CT, Souza CP, Soares D. Possible competitive displacement of Planorbids by Melanoides tuberculata in Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 2001;96:173-176.
- Paraense WL, Correa LR, Differential susceptibility of Biomphalaria tenagophila populations to infection with a strain of Schistosoma mansoni. J Parasitol. 1978;64:822-826.
- Martins Souza RL, Pereira CA, Coelho PM, et al. Silica treatment increases the susceptibility of the Cabo Frio strain of Biomphalaria tenagophila to Schistosoma mansoni infection but does not alter the natural resistance of the Taim strain. Parasitol Res. 2003;91:500-507.
- Rosa FM, Godard ALB, Azevedo V, et al. Biomphalaria tenagophila: dominant character of the resistance to Schistosoma mansoni and descendants of cross-breeding between resistant (Taim RS) and susceptible (Joinville SC) strains. Mem Inst Oswaldo Cruz. 2005;100:19-23.
- Marques DP, Rosa FM, Maciel E, et al. Reduced susceptibility of a Biomphalaria tenagophila population to Schistosoma mansoni after introducing the resistant Taim/RS strain of B. tenagophila into Herivelton Martins stream. PLoS One. 2014;9:e99573.
- Stothard JR, Llewellyn-Hughes J, Griffin CE, et al. Identification of snails within the Bulinus africanas group from East Africa by multiplex SNaPshotTM analysis of single nucleotide polymorphisms within the cytochrome oxidase subunit 1. Mem Inst Oswaldo Cruz. 2002;97:31-36.
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783-791.
- Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585-595.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular evolutionary genetics analysis. Mol Biol Evol. 2013;30:2725-2729.
- Bergquist NR, Colley DG. Schistosomiasis vaccine: Research to development. Parasitol Today. 1998;14:99-104.
- You H, Liu C, Du X, et al. Acetylcholinesterase and nicotinic acetylcholine receptors in schistosomes and other parasitic helminths. Molecules. 2017;22:1550.
- Campbell SJ, Biritwum NK, Woods G, et al. Tailoring water, sanitation, and hygiene (WASH) targets for soil-transmitted helminthiasis and schistosomiasis control. Trends Parasitol. 2018;34:53-63.
- Zahoor Z, Lockyer AE, Davies AJ, et al. Differences in the gene expression profiles of haemocytes from schistosome- susceptible and -resistant Biomphalaria glabrata exposed to Schistosoma mansoni excretory-secretory products. PLoS One. 2014;9:e93215.
- Plows LD, Cook RT, Davies AJ, et al. Phagocytosis by Lymnaea stagnalis haemocytes: A potential role for phosphatidylinositol 3-kinase but not protein kinase A. J Inver Pathol. 2006;91:74-77.
- Coelho PMZ, Carvalho OS, Andrade ZA, et al. Biomphalaria tenagophila/Schistosoma mansoni interaction: Premises for a new approach to biological control of schistosomiasis. Mem Inst Oswaldo Cruz. 2004;99:109-111.
- Frandsen F. Studies of the relationship between schistosoma and their intermediate hosts III, The genus Biomphalaria and Schistosoma mansoni from Egypt, Kenya, Sudan, Uganda, West Indies (St. Lucia) and Zaire (two different strains: Katanga and Kinshasa). J Helminthol. 1979;53:321-348.
- Lu L, Zhang S, Mutuku SM, et al. Relative compatibility of Schistosoma mansoni with Biomphalaria sudanica and B. pfeifferi from Kenya as assessed by PCR amplification of the S. mansoni ND5 gene in conjunction with traditional methods. Parasites Vectors. 2016;9:166.
- Webster BL, Rollinson D, Stothard JR, et al. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. J Helminthol. 2010;84:107-114.
- Stothard JR, Campbell SJ, Osei-Atweneboana MY, et al. Towards interruption of schistosomiasis transmission in sub-Saharan Africa: developing an appropriate environmental surveillance framework to guide and to support 'end game' interventions. Infect Dis Poverty. 2017; 6:10.
- Hanington PC, Forys MA, Loker ES. A somatically diversified defense factor, FREP3, Is a determinant of snail resistance to schistosome infection. PLoS Negl Trop Dis. 2012;6: e1591.
- Pila EA, Gordy MA, Phillips VK, et al. Endogenous growth factor stimulation of hemocyte proliferation induces resistance to Schistosoma mansoni challenge in the snail host. Proc Natl Acad Sci. 2016;113:5305-5310.
- Rollinson D, Stothard JR, Jones CS, et al. Molecular characterization of Intermediate snail hosts and the search for resistance genes. Mem Inst Oswaldo Cruz. 1998;93:111-116.
- Dejong RJ, Morgan JA, Wilson WD, et al. Phylogeography of Biomphalaria glabrata and B. pfeifferi, important intermediate hosts of Schistosoma mansoni in the new and old world tropics. Mol Ecol. 2003;12:3041-3056.
- Standley CJ, Wade CM. The population genetic structure of Biomphalaria choanomphala in Lake Victoria, East Africa: implications for schistosomiasis transmission. Parasites Vectors. 2014;7:524.
- Daniels SR, Phiri EE, Klaus S, et al.Multilocus Phylogeny of the afrotropical freshwater crab fauna reveals historical drainage connectivity and transoceanic dispersal since the Eocene.Syst Biol. 2015;64:549-567.
- Schulthei R, Bocxlaer B, Riedel F, et al.Disjunct distributions of freshwater snails testify to a central role of the Congo system in shaping biogeographical patterns in Africa. Evol Biol. 2014;14:42.