Original Article
, Volume: 16( 4) DOI: 10.21767/0972-768X.1000289Unprecedented Example of Metal Complexes With 1-Aminomethyl-1-Cyclohexane Acetic Acid
- *Correspondence:
- Amshumali MK , Department of Chemistry, Vijayanagara Sri Krishnadevaraya University, Jnana Sagara Campus, Cantonment, Vinayakanagar, Bellary, Karnataka, India, Tel: +91-9481968706; E-mail: amshumali.m@gmail.com
Received: October 15, 2018; Accepted: October 26, 2018; Published: October 30, 2018
Citation: Amshumali MK, Abhilash N, Patil M. Unprecedented Example of Metal Complexes With 1-Aminomethyl-1-Cyclohexane Acetic Acid. Int J Chem Sci. 2018;16(4):289
Abstract
A new coordination complexes of cobalt and nickel are synthesized using Gabapentin, [1-(amino methyl)-1-cyclohexane acetic acid] as a ligand. Complexes have been characterized by XRD. single crystal x-ray diffraction studies reveal that the cobalt complex (1) is coordinated with four gabapentin molecules and nickel complex (2) is with one gabapentin. Further, the ligand is in the neutral form in the complex (1) and in zwitter ion form in complex (2). Molecular docking study of complex (1) reveals that this complex may have better activity than gabapentin.
Keywords
Gabapentin; Molecular docking; Metal-gabapentin complex
Introduction
1-(amino methyl)-1-cyclohexane acetic acid commonly known as gabapentin is a neuroleptic drug that is usually prescribed in combination with other medication for the prevention of seizure disorders and for the treatment of some mood disorders, anxiety and tardive dyskinesia [1]. More recently gabapentin has been advanced for the treatment of neuropathic pain [2] however, the mechanism of action of gabapentin remains a subject of considerable discussion [3] many papers have been recently published on conformational studies of gabapentin-containing peptides [4], on stereochemistry [5] and on cyclization reactions involving gabapentin [6-10] and transition metal (Ru, Os, and Ir) catalysts. However, no coordination compounds of gabapentin with biologically active metal complexes have been reported except one in recent communication reported by Braga et.al [11] in which the mechanochemical preparations of zinc and copper complexes of gabapentin were reported. Since the knowledge on how the gabapentin molecule binds with the metal ions in the biological system is important to understand the mechanism of action of the gabapentin, we started the invitro synthesis and study of the gabapentin metal complexes. In this paper, we report the efficient and simple solution phase synthesis and structural characterization of gabapentin complexes with cobalt and nickel as metals for coordination. The molecular docking study on gabapentin-cobalt complex is also described in the present paper. The gabapentin complexes with cobalt and nickel can be easily achieved in the laboratory by heating the Stoichiometric quantity of metal salts (cobalt or nickel) with gabapentin, in water. Concentrating and cooling gave a precipitate of the resulting complex 1 and 2.
Experimental
Synthesis of cobal[gabapentin]4; {Co(H2O)2[gabapentin]4}
0.8 g (4.66 mmol) of gabapentin and 0.34 g (1.167 mmol) of cobaltous nitrate were taken in a 100 mL beaker and 50 mL of water was added, heated the mixture and reduced the volume to 20 mL, cooled in a refrigerator. Fine red color crystals start forming after 3 days. Crystals were separated by filtration. Yield: 65%. Elemental Analysis: Calc. C:46.03, H: 4.26, N:7.96% Found. C: 46.11, H: 4.22, N: 8.01%.
Synthesis of {Nickel (H2O)5[Gabapentin] SO4}
0.6g NiSO4.6H2O (2.326 mmol) and 0.8 g of gabapentin (2.326 mmol) were taken in a 100 mL beaker and 50 mL of water was added, heated the mixture and reduced the volume to 20 ml, cooled in a refrigerator. Fine green color crystals start forming after 3 days. Crystals were separated by filtration. Yield: 60%. Elemental Analysis: Calc. C: 29.95, H: 6.49, S:7.78 N: 3.36% Found: C: 29.87, H: 6.49, S: 7.84, N: 3.38%.
Results and Discussion
Crystal structures of 1 and 2
Single crystal formation of 1 and 2 was achieved by dissolving the complex in a minimum amount of water and subject to the slow evaporation in the laboratory conditions. A crystal of suitable size was mounted using a Mitigen micromount, and single-crystal data collection was done on a Bruker Smart X2S bench-top diffractometer equipped with a micro-focus MoKα radiation (λ=0.71073) at room temperature [8]. The machine was operated at 50 kV and 1 mA. Data reduction was performed using SAINTPLUS. Scaling, absorption correction was done using SADABS, all embedded in the Apex 2 software suite [8]. The crystal structure was solved by direct methods using XS and the structure refinement was done using XLin the SHELXTL package [9]. The position and thermal parameters of all the non-hydrogen atoms were refined. The hydrogens were fixed in geometrically calculated positions and refined isotropically. The ORTEP diagrams and packing diagrams were created using Mercury 3.0 and POV ray. Crystal and experimental data for compound 1 and 2 are listed in TABLE 1 and 2 respectively. Powder data were collected with para analytical x’pert pro equipped with x’Celerator detector. A Cu anode was used as X-ray source at 40 kV and 40 mA. The programme powder cell-2.2 was used for calculation of X-ray powder patterns.
| Compound identification | Red crystal |
|---|---|
| Chemical formula | CoC27H30N4O10 |
| Formula mass | 703.80 |
| Crystal system | Triclinic |
| Space group | P-1 |
| a (Å) | 12.5889 (6) |
| b (Å) | 12.6216 (5) |
| c (Å) | 14.3626 (7) |
| β (Å) | 100.466 (4) |
| Unit cell volume (Å3) | 2197.02 |
| Z | 15 |
| Temperature (K) | 296 (2) |
| ρcalc (g cm-3) | 1.172 |
| F(000) | 1840 |
| Crystal size (mm3) | 0.30 × 0.27 × 0.19 |
| No. of reflections measured | 4229 |
| No. of independent reflections | 2717 |
| Rint | 0.0571 |
| _ρmin, max (e Å-3) | -0.32, 0.24 |
| Final R1 values (I>2σ(I)) | 0.054 |
| Final wR(F2) values (I>2σ(I)) | 0.143 |
| Final R1 values (all data) | 0.098 |
| Final wR(F2) values (all data) | 0.167 |
| Goodness of Fit (GOOF) | 1.012 |
Table 1: Structural information of complex 1.
| Compound identification | Green crystal |
|---|---|
| Chemical formula | C9H27NSO11Ni |
| Formula mass | 416.09 |
| Crystal system | Triclinic |
| Space group | P-1 |
| a (Å) | 6.0146 (4) |
| b (Å) | 8.5196 (4) |
| c (Å) | 35.0416 (19) |
| β (Å) | 93.688(4) |
| Unit cell volume (Å3) | 1791 |
| Z | 0 |
| Temperature (K) | 296 (2) |
| ρcalc (g cm-3) | 1.693 |
| F(000) | 1840 |
| Crystal size (mm3) | 0.30 × 0.24 × 0.19 |
| No. of reflections measured | 2717 |
| No. of independent reflections | 2717 |
| Rint | 0.0622 |
| _ρmin, max (e Å-3) | -0.32, 0.24 |
| Final R1 values (I>2σ(I)) | 0.0711 |
| Final wR(F2) values (I>2σ(I)) | 0.143 |
| Final R1 values (all data) | 0.096 |
| Final wR(F2) values (all data) | 0.161 |
| Goodness of Fit (GOOF) | 1.078 |
Table 2: Structural information of complex 2.
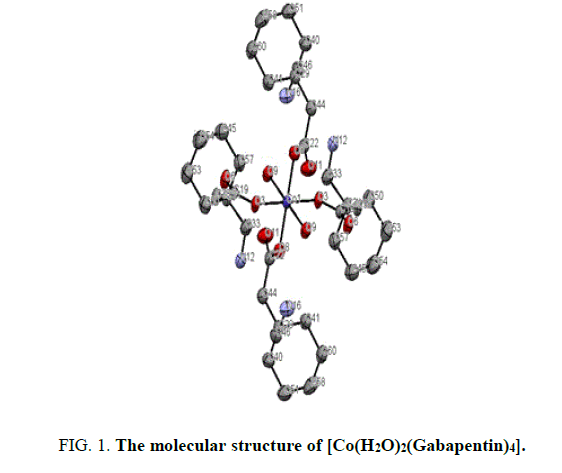
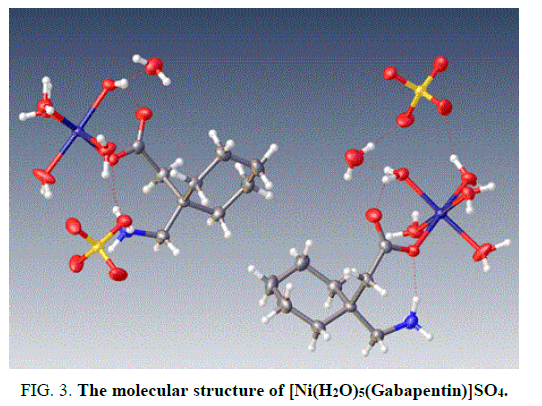
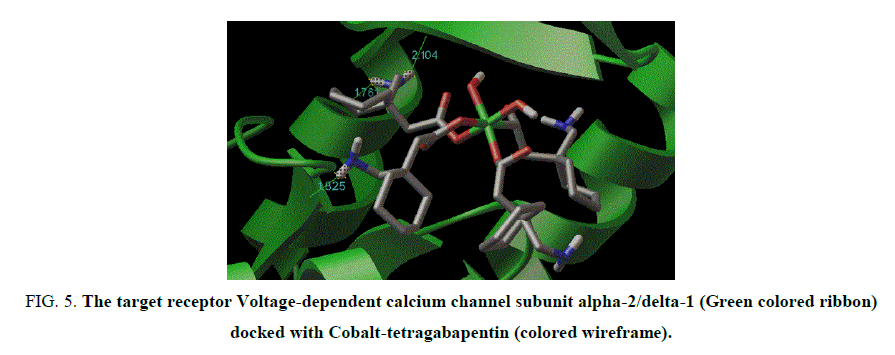
The X-ray structure determination allowed us for the unambiguous identification of the formation of the complex1. (FIG. 1). In which the Cobalt is octahedrally coordinated with two water molecule and four gabapentin molecules. The gabapentin coordinated via carboxylate oxygen with the Co-O bond length 2.064(2) A° and 2.088(2) A° all the bond lengths are within the expected range (FIG. 2). The detailed structural information is given in the TABLE 1. In the complex 2, (FIG. 3-7) the gabapentin present in the zwitter ion form and the corboxylate oxygen is binds to nickel with the bond length Ni.....O is about 2.036 A°. The detailed structural information is given in the TABLE 2.
Figure 5: The target receptor Voltage-dependent calcium channel subunit alpha-2/delta-1 (Green colored ribbon) docked with Cobalt-tetragabapentin (colored wireframe).
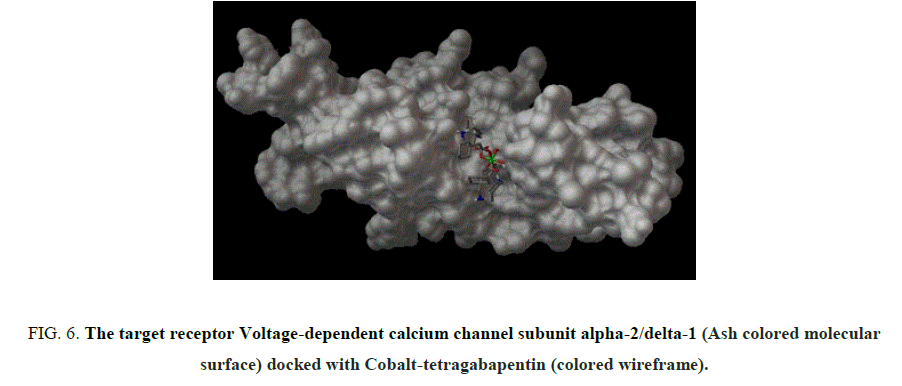
Figure 6: The target receptor Voltage-dependent calcium channel subunit alpha-2/delta-1 (Ash colored molecular surface) docked with Cobalt-tetragabapentin (colored wireframe).
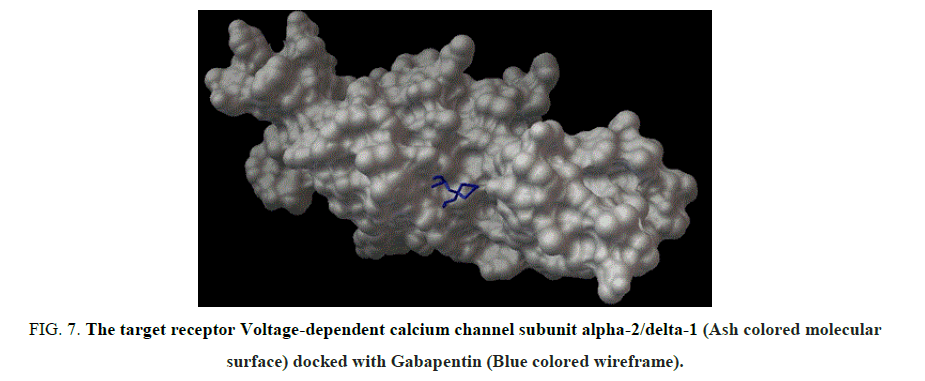
Figure 7: The target receptor Voltage-dependent calcium channel subunit alpha-2/delta-1 (Ash colored molecular surface) docked with Gabapentin (Blue colored wireframe).
Molecular Docking of complex 1
In silco study: Chem-Sketch of ACDLABS 10.00 software was used to design the ligands Cobalt-tetra-gabapentin and gabapentin followed by 3D optimization. The Sybyl Mol2 format files of these ligands were converted into Protein data bank (pdb) format using Open Babel software [10]. The Gabapentin target receptor Voltage-dependent calcium channel subunit alpha-2/delta-1 and its amino acid sequence were elicited from Drugbank [11]. A 3D model of the amino acid sequence of target protein was built by ESyPred3D web server [12] using the 3D structure PDB Id: 3LIA chain 'A' as template. Gastieger atom charges, solvent deletion and hydrogens were added into the receptors files for the preparation of receptor in docking simulation by Autodock 4.2 [13]. Docking analysis was done using Autodock 4.2 [13-17]. Torsions in ligands were set to 6 and non-polar hydrogens present in receptors were merged. AutoGrid 4.2 program, supplied with AutoDock 4.2 was used to produce grid maps. The grid box size was set at 62, 62 and 62 A° (x, y, and z) to include all the amino acid residues present in the active site of macromolecule. The Lamarckian Genetic Algorithm (LGA) was chosen for the search of best conformers with lowest binding energy. Results were analyzed to study the interactions, binding energy and hydrogen bond interactions for the best conformers and given in the TABLE 3.
| Parameters | Gabapentin | Cobalt-tetragabapentin |
|---|---|---|
| Binding energy | -5.5 | -7.64 |
| Ligand efficiency | -0.46 | -0.15 |
| Inhibition Constant | 92.71 µM | 2.5 µM |
| Intermolecular energy | -7.29 | -15.1 |
| Electrostatic energy | -2.72 | -5.69 |
| Vdw_hb_desolvation energy | -4.57 | -9.4 |
| Total internal energy | 0.05 | 0.31 |
| Torsional energy | 1.79 | 7.46 |
| Unbound energy | 0.05 | 0.31 |
| H-bonds | 03 | 03 |
Table 3: Comparative molecular docking results of Gabapentin and Cobalt-tetragabapentin generated from Autodock 4.2.
This result clearly indicates that the complex 1 may have better efficiency over gabapentin alone.
Conclusion
For the first time we have studied the gabapentin complexes that contain cobalt and nickel metal ions. Molecular docking studies shows the complexes are more active than the gabapentin alone. Further, in vivo studies of the complexes to prove the efficacy and safety as a drug are underway.
Acknowledgements
Authors are grateful to Prof. Guru Row, SSCU, IISc, Bangalore for providing the instrumental facilities and for VSK University for the research facilities.
References
- Jensen AA, Mosbacher J, Elg S. The anticonvulsant gabapentin (Neurontin) does not act through γ-aminobutyric acid-B receptors. Mol Pharmacol. 2002;61:1377.
- Taylor CP. Emerging perspectives on the mechanism of action of gabapentin. Neurology. 1994;44:S10-6.
- Sutton KG, Snutch TP. Gabapentin: A novel analgesic targeting voltage‐gated calcium channels. Drug Development Res. 2001;4(3):167-72.
- Taylor CP, Gee NS, Su TZ. A summary of mechanistic hypotheses of gabapentin pharmacology. Epilepsi Res. 1998;29:233.
- Eisenberg E, River Y, Shifrin A, et al. Antiepileptic drugs in the treatment of neuropathic pain. Drugs. 2007;67(9):1265-89.
- Rosenberg JM, Harrell C, Ristic H, et al. The effect of gabapentin on neuropathic pain. The Clinical J of Pain. 1997;13(3):251-5.
- Maneuf YP, Gonzalez MI, Sutton KS, et al. Cellular and molecular action of the putative GABA-mimetic, gabapentin. Cellular and Mol Life Sci CMLS. 2003;60(4):742-50.
- Vasudev PG, Shamala N, Ananda K, et al. C9 Helices and ribbons in γ‐peptides: crystal structures of gabapentin oligomers. Angewandte Chemie. 2005;117(31):5052-5.
- Ananda K, Aravinda S, Vasudev PG. Stereochemistry of gabapentin and several derivatives: solid state conformations and solution equilibria. Curr Sci. 2003; 85:1002.
- Bryans JS, Chessum NE, Huther N. Metal-catalysed radical cyclisations leading to N-heterocycles: new approaches to gabapentin and pulchellalactam. Tetrahedron. 2003;59(33):6221-31.
- Braga D, Grepioni F, Maini L, et al. Simple and quantitative mechanochemical preparation of the first zinc and copper complexes of the neuroleptic drug gabapentin. Cryst Eng Comm. 2008;10(5):469-71.
- Bruker A. APEX2, GIS, SADABS and SAINT; Bruker AXS Inc: Madison, Wisconsin, USA. 2009.
- Sheldrick GM. A Short History of SHELX. Acta Crystallographica Section A: Foundations of Crystallography. Acta Crystallogr A. 2008;64:112-22.
- M O’Boyle N, Banck M, James CA. Open Babel: An open chemical toolbox. J Cheminformatics. 2011;3:33.
- Goto M, Miyahara I, Hirotsu K. Structural determinants for branched-chain aminotransferase isozyme specific inhibition by the anticonvulsant drug gabapentin. J Biol Chem. 2005;280:37246-56.
- Pettersen EF, Goddard TD, Huang CC. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605-12.
- Sanner MF. Python: a programming language for software integration and development. J Mol Graphics Mod. 1999;17:57-61.