Original Article
, Volume: 9( 5)Fluorimetric Study of the Interaction of Daidzein with BSA
Song J1, Yang L1,2 and Wang QM1*
1School of Pharmacy, Jiangsu Provincial Key Laboratory of Coastal Wetland Bio Resources and Environmental Protection, Yancheng Teachers’ University, Yancheng, Jiangsu 224051, People’s Republic of China
2Institute of Nautical Medicine, Nantong University, Nantong, Jiangsu 224051, People’s Republic of China
- *Correspondence:
- Wang QM, School of Pharmacy, Jiangsu Provincial Key Laboratory of Coastal Wetland Bio Resources and Environmental Protection, Yancheng Teachers’ University, Yancheng, Jiangsu 224051, People’s Republic of China, Tel: 0086-18571714629, E-mail: wang0qingming@163.com
Received: June 04, 2016; Accepted: July 02, 2016; Published: July 08, 2016
Citation: Song J, Yang L, Wang QM. Fluorimetric Study of the Interaction of Daidzein with BSA. ChemXpress. 2016;9(5): 104.
Abstract
The mutual interaction of daidzein with bovine serum albumin (BSA) was investigated by means of fluorescence spectroscopy. The results revealed that daidzein caused the fluorescence quenching of BSA through a static quenching procedure. The value of binding constant (Ka) were 1.17 × 104 M−1 (T=310K), 3.74 × 104 M−1 (T=303K), 5.36 × 104 M−1 (T=298K) respectively. Thermodynamic parameters ΔG0, ΔH0, ΔS0 were calculated, besides the van der Waals force and hydrogen bonds play a significant role in spontaneous stabilizing the complex. Moreover, the effect of Cu2+, Ni2+, Mn2+ and Co2+ on the binding constants between daidzein and BSA were studied.
Keywords
Daidzein; Bovine serum albumin (BSA); Fluorescence quenching; Binding constant
Introduction
Daidzein (Figure 1) produced during the growth of leguminous plants such as soybean is a sort of secondary metabolite with high nutritional value. Researches have revealed that daidzein possesses diverse biological functions. Daidzein could bind to estrogen receptors on bone cells, studied by Peterson et al., which can reduce the incidence of osteoporosis effectively [1]. Besides, Ward et al. reported that daidzein could lower incidence of colorectal cancer of females and prostate cancer of males [2]. According to the discussion above, the medicinal properties of daidzein should be paid more attention to.
Serum albumin is a kind of vital plasma protein distributing in the blood, and own multiple biological functions [3-5], such as metabolites transport and drug delivery [6]. Therefore, bovine serum albumin is the optimum selection to be the target protein for reaching drug efficacy. In recent years, many findings were reported in this field [7,8].
In this paper, the fluorescent spectrometry was employed to study the mechanism of mutual interaction between daidzein and bovine serum albumin (BSA) for the first time. The binding constant, binding sites and thermodynamic parameters, which are obtained from spectroscopic data, can provide the information and evidence for micro-molecule drugs binding to BSA, a polymer carrier.
Experimental
Experimental material
BSA was purchased from Sino-Biotechnology Company (Shanghai, China). Daidzein was purchased from Sigma (USA), the purity of which is not less than 98%. The buffer Tris-HCl, NaCl, HCl were purchased commercially and used without further purification.
Physical material
Fluorescence emission spectra were recorded on RF-5301 Spectrophotometer (Shimadzu, Japan) with 1.0 cm quartz cells. The emission and excitation slits were 3 nm and 3 nm, respectively. Fluorescence quenching spectra were measured in the range of 300 nm to 500 nm with the excitation wavelength of 285 nm at three temperatures (298 K, 303 K and 310 K). The pH value of Tris-HCl was measured using a pH-2500 pH-meter.
Fluorescence spectrometry
The daidzein (0.0024 g) was dissolved in DMSO to 1.0 × 10-2 M, and then the prepared solution was diluted to 1 × 10-3 M and 1 × 10-4 M. BSA (0.0068 g) was dissolved in Tris-HCl (25 mM, pH=7.40) buffer solution to 1.0 × 10-2 M as stock solution, afterwards the obtained solution was diluted to 1 × 10-3 M and 1 × 10-4 M for further use.
For titration experiments, 5 μL daidzein solution (1.0 × 10-4 M) was added to a solution, which containing appropriate concentration of BSA is 1.0 × 10-6 M (2 mL, in 25 mM Tris-HCl, pH 7.40). The resulting solution was left to react for 3 min at three temperatures (298 K, 303 K and 310 K). The emission and excitation slits were 3 nm and 3 nm, respectively. Fluorescence spectra were measured in the range of 300 nm to 500 nm at the excitation wavelength of 285 nm and the emission wavelength of 345 nm.
Results and Discussion
Daidzein fluorescence spectra of BSA
Fluorescence spectroscopy is an effective method to study the interaction between micro-molecules and bio-molecules, and the binding constants can be calculated according to experimental data. The BSA molecule, containing tryptophan, tyrosine and phenylalanine residues, processes the intrinsic fluorescence, meanwhile daidzein was found without fluorescence. Thus the fluorescence spectroscopy was used to study the interaction between daidzein and BSA, the binding constants, binding sites at different temperature were measured by fluorescence spectra.
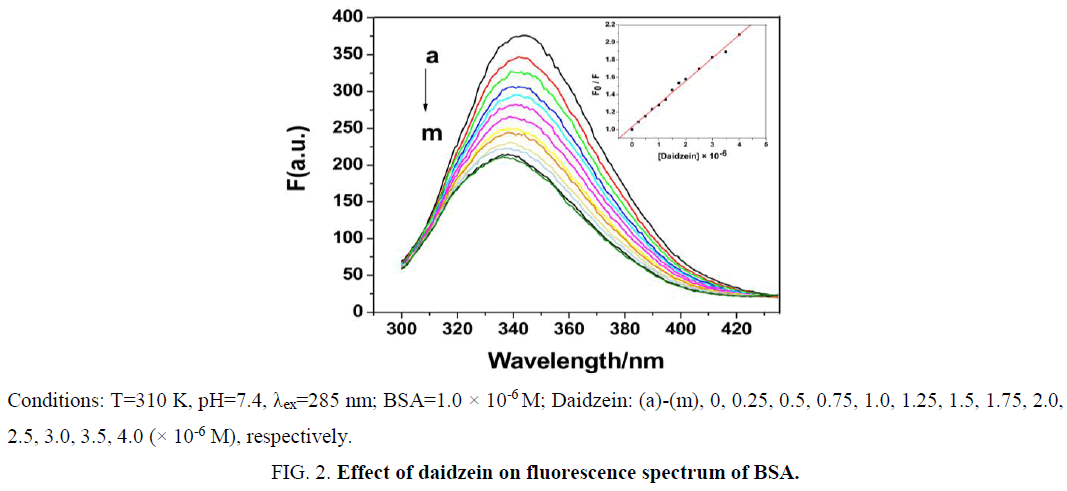
The effect of daidzein on BSA fluorescence intensity is shown in Figure 2. With gradual addition of daidzein into BSA solution at 310 K, the fluorescence intensity of BSA at 345 nm remarkably decreased, which suggested the interaction between daidzein and BSA. The fluorescence of BSA was quenched by daidzein may be caused by excited-state reactions, molecular rearrangements, energy transfer, ground-state complex formation or collisional quenching [9].
The mechanism fluorescence quenching
The mechanism of fluorescence quenching is generally divided into static quenching and dynamic quenching. If the interaction between daidzein and BSA is static quenching, the fluorescence quenching in the daidzein and BSA system could be described by Stern-Volmer equation [10]:
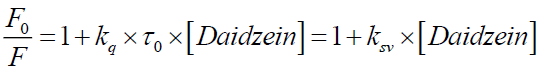 (1)
(1)
Where, F0 and F are the fluorescence intensities of BSA in Tris-HCl (25 mM, pH=7.40) buffer solution in the absence and presence of daidzein, respectively; kq is the quenching rate constant; ksv is the Stern-Volmer quenching constant; τ0 is the average lifetime of the protein in the absence of a quencher, τ0 is generally about 10-8 s for a biopolyme; [Daidzein] is the molar concentration of the respective quencher.
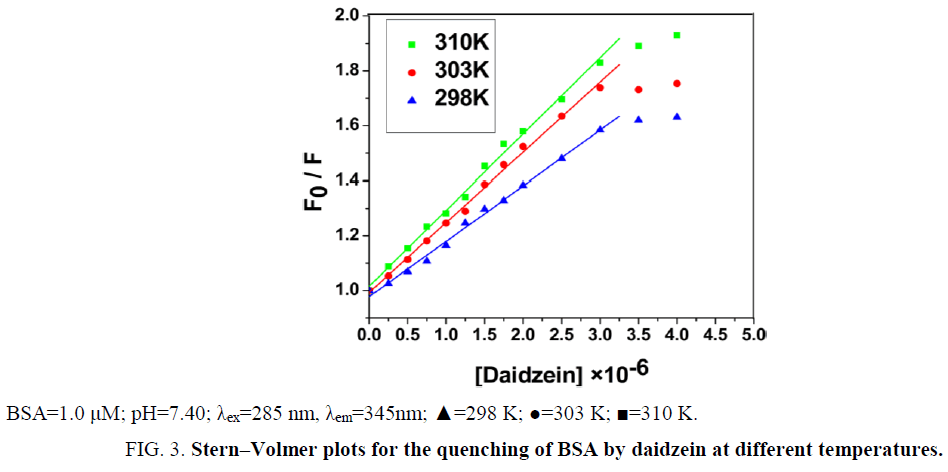
From Eq. (1), ksv can be determine by linear regression of a plot of F0/F against [Daidzein]. As shown in Figure 3, a good linearity of F0/F versus [Daidzein] was found when the concentration of daidzein is 0 μM to 3.0 μM. According to kq=ksv/τ0 [11], the kq (the values of ksv and kq were illustrated in Table 1) is much greater than 2.0 × 1010 M-1 s-1, indicating that the quenching mechanism of BSA by daidzein was not initiated by dynamic collision but by static quenching interaction [12]. Besides, the values of ksv decreased with the rising temperature at low concentrations of daidzein, suggesting that the probable quenching mechanism of interaction was initiated by formation of a ground-state complex [13,14].
| pH | T(K) | ksv±SD(104L.mol-1) | kq±SD(1012L.mol-1.S-1) | R |
|---|---|---|---|---|
| 7.40 | 298 | 7.95 ± 0.009 | 7.95 ± 0.009 | 0.997 |
| 303 | 5.30 ± 0.008 | 5.30 ± 0.008 | 0.993 | |
| 310 | 3.19 ± 0.004 | 3.19 ± 0.004 | 0.994 |
Table 1. Binding and quenching constants according to Stern-Volmer curves.
The binding constant and binding sites
It is known to all that there are independent binding sites in the biomolecule, and the binding constant Ka, as well as number of binding site n could be calculated by using the double logarithm equation[15,16]:
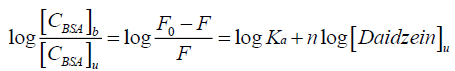 (2)
(2)
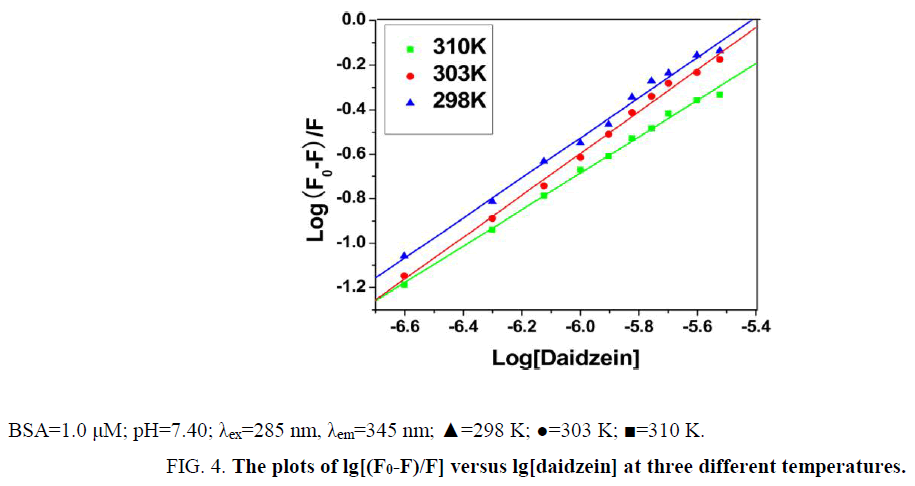
Where [CBSA]b is molar concentration of BSA with a quencher bound and [CBSA]u is molar concentration of free BSA (with no quencher bound), F0 and F for fluorescence intensities of BSA in the absence and presence of daidzein, Ka for the cumulative binding constant, n for the number of binding sites, and [Daidzein]u for the molar concentration of free daidzein. Figure 4 shows the plots of log [(F0-F)/F] versus log [Daidzein] for the daidzein-BSA system at different temperatures, and the calculated results are presented in Table 2. As shown in Table 2, there is only a single class of binding sites on BSA and daidzein, and the binding constant (Ka) decreased with rising temperature because the stability of the complex reduced with a rise in temperature. Binding constant decreases with increasing temperature, which further confirmed is that a static quenching process between daidzein and BSA [17].
| T | Ka ± SD(104L.mol-1) | n ± SD | R |
|---|---|---|---|
| 298 | 5.36 ± 0.028 | 0.98 ± 0.011 | 0.996 |
| 303 | 3.74 ± 0.011 | 1.02 ± 0.024 | 0.998 |
| 310 | 1.17 ± 0.015 | 0.95 ± 0.008 | 0.994 |
Table 2. The binding constants and binding sits of between daidzein and BSA at pH 7.4.
Thermodynamic analysis of the interaction between daidzein and BSA
The van der Walls interaction, hydrogen bonds, electrostatic and hydrophobic interactions were found between protein interactions with small molecular substrates [18]. The thermodynamic parameters for protein reactions can be accounted for the main forces contributing to protein stability. If the enthalpies change (ΔH) does not vary significantly over the temperature range studied, then its value and that of entropy change (ΔS) can be determined from the van’t Hoff equations [19]:
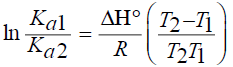 (3)
(3)
Gibbs free energy change (ΔG) can be obtained from
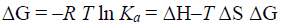 (4)
(4)
Where R is the gas constant.
By the function Eq. (3) and (4), thermodynamic parameters ΔH, ΔS and ΔG of the interaction between daidzein and BSA could be obtained, the results were shown in Table 3. The findings of Ross [20] and the values in Table 3 indicated that the process of interaction between daidzein and BSA is spontaneous and that the main binding forces are hydrogen bonds and van der Waals forces.
| T | ΔH° (KJ.mol-1) | ΔGo (KJ.mol-1) | ΔSo (KJ.mol-1.K-1) |
|---|---|---|---|
| 298 | -58.78 | -27.96 | -0.10 |
| 303 | -27.41 | ||
| 310 | -26.73 |
Table 3. The thermodynamic parameters between daidzein and BSA at pH=7.40.
The fitted curve of ln Ka versus 1/T was shown in Figure 5. As shown in Figure 5, the enthalpy change (ΔH) is calculated from the slope of the van’t Hoff relationship and the free energy change (ΔG) is estimated from Eq. (4) and summarized in Table 3. The negative values of ΔG indicates that the binding process is spontaneous, while a positive ΔS value is frequently taken as typical evidence for hydrophobic interactions and negative ΔH values indicate that hydrogen bonds and van der Waals force play a part in the binding [21].
Effect of metal ions on the interaction between daidzein and BSA
The effects of metal ions M2+ (M=Cu, Co, Mn, Ni) on the interaction between daidzein and BSA was solidified by fluorescence spectroscopy (Figure 6). The M2+ was added to the BSA solution at the temperature of 310 K forming complexes with molar ratio of 1:1 (the concentration of BSA and metal ion was 1.0 × 10-6 M). Then the fluorescence spectra of the interaction between daidzein and BSA-M2+ system were measured in 25 mM, pH=7.40, Tris-HCl buffer solution. The results shown that the fluorescence intensity of BSA-M2+ system in 345 nm was decreased with the addition of daidzein until the equilibrium was achieved, this indicated that the endogenous fluorescence of BSA was quenched by the addition of daidzein at the presence of M2+. According to the Eq. (2), the binding constant of the interaction between daidzein and BSA can be calculated when the metal ions exist, and the results are shown in Table 4. As shown in Table 4, the presence of Mn2+, Co2+ and Cu2+ led the value of Ka of the interaction between BSA and daidzein increased, which could cause by that M2+ first formed a complex with daidzein, and then combined with BSA to form a ternary complex. The results are similar to our early research [22,23]. The addition of M2+ made the free concentration of daidzein decreased, made the retention time of daidzein in plasma to be prolonged, which indicates that the phenomenon could be helpful to the sustained release of drug [24].
| System | Ka ± SD(104L.mol-1) | n ± SD | r | Ka'/Ka |
|---|---|---|---|---|
| BSA-Daidzein | 3.19 ± 0.011 | 1.05 ± 0.011 | 0.997 | 1 |
| BSA- Daidzein - Cu2+ | 23.4 ± 0.013 | 0.92 ± 0.013 | 0.998 | 7.33 |
| BSA- Daidzein - Ni2+ | 1.32 ± 0.008 | 1.03 ± 0.014 | 0.997 | 0.41 |
| BSA- Daidzein -Mn2+ | 5.31 ± 0.005 | 1.02 ± 0.004 | 0.997 | 1.66 |
| BSA- Daidzein - Co2+ | 10.1 ± 0.022 | 1.12 ± 0.008 | 0.997 | 3.17 |
Table 4. The effect and metal ions on the interaction between daidzein and BSA at 310 K.
Conclusions
In this paper, the interaction between daidzein and bovine serum albumin (BSA) at different temperatures was studied by fluorescence spectra. The results showed that daidzein could cause static quenching BSA. Daidzein and BSA could form complexes with molar ratio of 1:1 at a certain temperature, and the binding constants were 1.17 × 104 M-1 (310 K), 3.74 × 104 M-1 (303 K) and 5.36 × 104 M-1 (298 K). Thermodynamic study results show that, if ΔG<0, the process of interaction between daidzein and BSA is spontaneous, and the value of ΔS and ΔH were less than 0. All the results reveal that the hydrogen bonding and van der Waals forces are the main forces in this process. The association constant, binding potential point and binding site between BSA with daidzin was discussed. The binding of BSA and daidzin was strengthened in the presence of Cu2+, Ni2+, Mn2+, Co2+ and the binding constans augment 7.33, 0.41, 1.66 and 3.17, respectively. We believe that this work will help to enrich the knowledge on these intermolecular interactions, further study in this area, and assist to provide useful information of the structural features that determine the therapeutic effectiveness of drugs and design of dosage forms.
Acknowledgments
This work were financially supported by The National Natural Science Foundation for Young Scientists of China (21301150); The Natural Science Foundation of the Jiangsu Higher Education Institutions of China (13KJB150037); The Foundation of Jiangsu Provincial Key Laboratory of Coastal Wetland Bio Resources and Environmental Protection (JLCBE14007, JLCBE10006); The Foundation of Jiangsu Provincial Key Laboratory of Solonchak (JKLBS2012022); The Practice Innovation Training Program Projects for the Jiangsu College Students (201510324022Y); The Natural Science Foundation of Yancheng Teachers’ University (14YCKL006).
References
- Peterson CA, Schnell JD, Kubas KL, et al. Effects of soy isoflavone consumption on bone structure and milk mineral concentration in a rat model of lactation-associated bone loss. Eur J Nutr. 2009;48(2):84-91.
- Ward HA, Kuhnle GG, Mulligan AA, et al. Breast, colorectal, and prostate cancer risk in the European Prospective Investigation into Cancer and Nutrition-Norfolk in relation to phytoestrogen intake derived from an improved database. Am J Clin Nutr. 2010;91(2):440-8.
- Carter DC, Ho JX. Structure of serum albumin. Adv Protein Chem. 1994;45:153-203.
- Xiao Q, Huang S, Ma JQ, et al. JPhotochemPhotobiolA Chem.2012;249:5.
- Zhang Y, Qi Z, Zheng D, et al. Interactions of chromium (III) and chromium (VI) with bovine serum albumin studied by UV spectroscopy, circular dichroism, and fluorimetry. Biol Trace Elem Res. 2009;130(2):172-84.
- Ahmad B, Parveen S, Khan RH. Effect of albumin conformation on the binding of ciprofloxacin to human serum albumin: a novel approach directly assigning binding site. Bio Macromol. 2006;7(4):1350-6.
- Yue HL, Hu YJ, Huang HG. Spectrochimica ActaAMolBiomolSpectrosc. 2004;130:40.
- Yu X, Liao Z, Yao Q, et al. Spectroscopic studies on the interaction of Phacolysin and bovine serum albumin. Spectrochim Acta A Mol Biomol Spectrosc. 2014;127:231-36.
- Yusakul G, Sakamoto S, Juengwatanatrakul T, et al. Preparation and application of a monoclonal antibody against the isoflavone glycoside daidzin using a mannich reaction-derived hapten conjugate. Phytochem Anal. 2016;27(1):81-8.
- Ding F, Zhao G, Huang J, et al. Fluorescence spectroscopic investigation of the interaction between chloramphenicol and lysozyme. Eur J Med Chem. 2009;44(10):4083-9.
- Sengupta B, Chakraborty S, Crawford M, et al. Characterization of diadzein-hemoglobin binding using optical spectroscopy and molecular dynamics simulations. Int J Biol Macromol. 2012;51(3):250-8.
- Annaraj B, Balakrishnan C, Neelakantan MA. Synthesis, structure information, DNA/BSA binding affinity and in vitro cytotoxic studies of mixed ligand copper (II) complexes containing a phenylalanine derivative and diimine co-ligands. J Photochem Photobiol B Biol. 2016;160:278-91.
- Orozco-Gonzalez Y, Bistafa C, Canuto S. Solvent effect on the Stokes shift and on the nonfluorescent decay of the daidzein molecular system. J Phys Chem A. 2013;117(21):4404-11.
- Wu QH, Wang DY, Zhou X, et al. Study on the interaction between daidzein and human serum albumin. Guang Pu Xue Yu Guang Pu Fen Xi. 2009;29(7):1911-4.
- Wang H, Zhang M, Lv Q, et al. Determination of berberine and the study of fluorescence quenching mechanism between berberine and enzyme-catalyzed product. Spectrochim Acta A Mol Biomol Spectrosc. 2009;73(4):682-6.
- Dasgupta N, Ranjan S, Patra D, et al. Bovine serum albumin interacts with silver nanoparticles with a "side-on" or "end on" conformation. Chem Biol Interact. 2016;253:100-11.
- Vaishanav SK, Korram J, Nagwanshi R, et al. Adsorption Kinetics and Binding Studies of Protein Quantum Dots Interaction: A Spectroscopic Approach. J Fluoresc. 2016;26(3):855-65.
- Wang Y, Zhang H, Kang Y, et al. Effects of perfluorooctane sulfonate on the conformation and activity of bovine serum albumin. J Photochem Photobiol B Biol. 2016;159:66-73.
- Sudha A, Srinivasan P, Thamilarasan V, et al. Exploring the binding mechanism of 5-hydroxy-3',4',7-trimethoxyflavone with bovine serum albumin: Spectroscopic and computational approach. Spectrochim Acta A Mol Biomol Spectrosc. 2016;157:170-81.
- Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20(11):3096-102.
- Wang Q, Huang CR, Jiang M, et al. Binding interaction of atorvastatin with bovine serum albumin: Spectroscopic methods and molecular docking. Spectrochim Acta A Mol Biomol Spectrosc. 2016;156:155-63.
- Wang QM, Liu J, Zhang TX, et al. Fluorimetric study the interaction between asiatic acid and bovine serumalbumin. ChemXpress. 2015;8(1):18-25.
- Wang QM, Liu J, Zhang TX, et al. The Investigation of the Interaction between Daidzin and Bovine Serum Albumin by Fluorescence Spectroscopy. ApplMechMater. 2014;66:402.
- Zhang W, Jiang P, Chen Y, et al. Nanoscale. 2016;8:957.