Research
, Volume: 15( 2) DOI: 10.21767/0974-7532.1000151DNA Barcoding and Sequence Annotation Study of Diplokhema butyracea Spp. (Chiuri) from Nepal
- *Correspondence:
- Deepak Sharma and Janardan Lamichhane
Department of Biotechnology
School of Science, Kathmandu University
Nepal
Tel: +977-9851249996
E-Mail: deepakshrm59@gmail.com/ljanardan@ku.edu.np
Received: April 30, 2020; Accepted: May 22, 2020; Published: May 29, 2020
Citation: Sharma D, Shrestha TM, Lamichhane J. DNA Barcoding and Sequence Annotation Study of Diplokhema butyracea Spp. (Chiuri) from Nepal. Res Rev Biosci. 2020;15(2):151.
Abstract
Diplokhema butyracea spp. (Chiuri), also known as the butter tree, is a Himalayan medicinal plant of Nepal with high economic value. The use of molecular genetic method of DNA barcoding for the authentication and studies of these plants in specific sites can have a positive impact on the use of raw materials from this plant in herbal medicinal products. The Internal Transcribed Spacer (ITS) gene locus been shown to be the most effective source of DNA barcodes for this plant. Using Basic Local Alignment Search Tool (BLAST) searching of GenBank and Neighbor Joining (NJ) analysis shows very close sequence similarities (99.4%) with the other species of Diplokhema (National Center for Biotechnology Information (NCBI) accession number MT497981) from other countries. As this medicinal plant has been used as an important herbal drug ingredient, the use of this marker gene (ITS) can allow researchers to identify material from even highly degraded DNA samples or in the adulterants from herbal products. Accurate identification of this material is especially important in the case of herbal preparations and for biologically active additives including studies of heavy metal or minerals in the future.
Keywords
DNA barcode; Omics; Mega X; Adulterants; Herbal products
Abbreviations
DNA: Deoxyribonucleic Acid; ITS: Internal Transcribed Spacer; BLAST: Basic Local Alignment Search Tool; NJ: Neighbor Joining; NCBI: National Center for Biotechnology Information; TLC: Thin-Layer Chromatography; HPLC-UV: High-Performance Liquid Chromatography-Ultra-Violet; HPLC-MS: High-Performance Liquid Chromatography-Mass Spectrometry; PCR: Polymerase Chain Reaction; CTAB: Cetyl Trimethyl Ammonium Bromide; RNase: Ribonuclease
Introduction
The deciduous tree Diplokhema butyracea spp. (Chiuri) belongs to the family Sapotaceae. This tree, typically about 20 m in height, is distributed throughout Nepal, mainly on open hillsides of 400 to 1,800 meters in elevation in the sub-Himalayan tracts, and in northern Nepal, India and Bhutan [1]. It is also known as the Nepalese butter tree, a Himalayan medicinal plant. The main product of the tree is known as Chiuri ghee, and this is ghee or fat extracted from the seeds. Nowadays it has also become an economically important plant due to honey cultivation and [2] for enhancing the economic growth of rural developments in western Nepal.
The overall objective of this project is to screen the economically important medicinal plants of Nepal [3], their bio prospecting approach [4] and authenticate their identification and taxonomic status using DNA barcoding and the Mega X software [5]. The butter tree described here will be included along with a phytochemical analysis, biological activities and chemical constituents analysis as studied in other similar plants in previous [6,7]. Previous studies have reported the triglyceride and fatty acid composition of phulwara butter collected from India [8,9], but to date no molecular studies have reported for the analysis of Chiuri plants from Nepalese sources. Hence, the present investigation was carried out to produce molecular data for DNA barcodes and for phylogenetic studies [10] of the distribution of Chiuri plants in Nepal using a bioinformatics approach.
Traditional methods of medicinal plant identification include organoleptic methods using the senses of taste, sight, smell, touch etc. as well as macroscopic and microscopic methods based on identification by shape, color, texture and chemical profiling methods such as Thin-Layer Chromatography (TLC), High-Performance Liquid Chromatography- Ultra-Violet (HPLC-UV), and High-Performance Liquid Chromatography-Mass Spectrometry (HPLC-MS). However, using macroscopic and microscopic examinations, no single method can easily distinguish closely related species. Even the methods based on chemical profiles and markers may be affected by physiological changes and storage conditions. Authentication at the DNA level provides more reliability as it is a stable macromolecule which is found in all tissues and will not affected by external factors. Therefore, development of DNA-based markers is important for authentication of medicinal plants.
A novel technique of identifying biological specimens using short DNA sequences from either nuclear or organelle genomes is called DNA barcoding. The use of the term ‘DNA barcode’ as a taxonomic identifier was first proposed by Paul Hebert of University of Guelph in 2003 [11]. In this research we found that ITS gene obtain derived from ribosomal gene yielded the single highest discrimination rate (99.08%), but due to difficulties in obtaining high-quality sequences from leafs and other parts of the plants such as Diplokhema butyracea spp, others have argued that the better overall choice for seems to be the standard barcode based on rbcL+matK sequences. Because of failures to amplify in Polymerase Chain Reaction (PCR) reactions, we chose to use ITS sequences as this amplified well and yielded discrimination rates close to 90% using the distance-based method Taxon DNA [12]. However, more complete specimen sampling may be needed to decide on the best analytic method.
Methodology
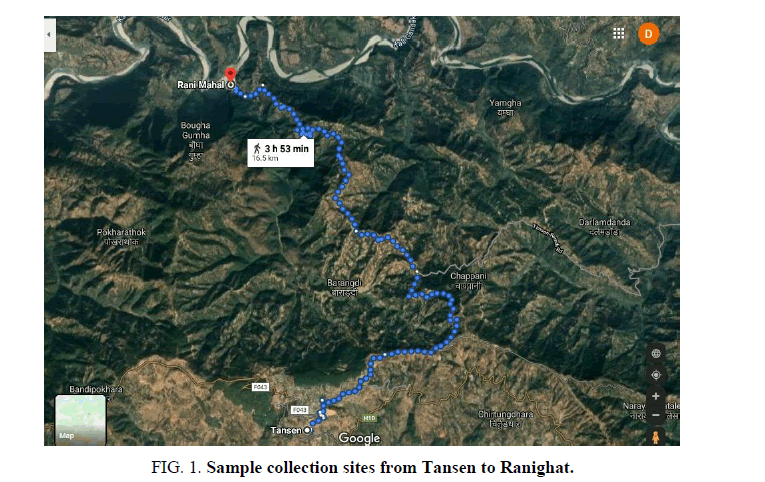
Random samples were collected from the natural habitat of the mid hill region of the Palpa district in the month of April 2017. Collections were obtained from 5 different sites in the Tansen to Ranighat area forest (FIG. 1). The samples were identified by a taxonomist of Kathmandu University faculty from the department of Pharmacy, Mrs. Tirtha Maiya Shrestha. Species materials were submitted to the herbarium to get an accession code which was later used for NCBI sequence submission (KU_2017_CHRi 01).
DNA extraction
Leaf material collected was used for the DNA extraction and analysis from 6 samples of Diploknema using some modifications of the Cetyl Trimethyl Ammonium Bromide (CTAB) method [13]. The recovery of DNA in mg/gm was calculated by measuring absorbance at 260 nm/290 nm. The ratio of absorbance 260 nm/280 nm was found to be in the range of 1.8 to 2.0, and the DNA yield ranged from 0.67 μg/ml to 0.86 μg/ml. This shows that the DNAs recovered were of high quality containing very low contamination of terpenoids and polysaccharides. The chemicals used in the isolation of DNA by the CTAB method increase DNA purity by removing all impurities. Long term chloroform isoamyl alcohol treatment removes chlorophyll, pigments and dyes. Overnight treatment using Ribonuclease (RNase) was also done to degrade RNA. Other potential contaminants (detergents, protein, polysaccharides etc.) were removed by an additional step of phenol: chloroform: isoamyl alcohol (25:24:1 v/v) and phenol: chloroform (24:1) extraction (FIG. 2).
PCR and sequencing
Genomic DNA extracted from all plant samples were quantified with spectrophotometric analysis and used for PCR with all the 3 gene loci for barcoding following iBOLD guidelines. Primers used to amplify and sequence cpITS2 and cpITS3 were those described earlier for the cpITS4 region: 5′?G TATTCTGGTGTCCTAGGCGTAG?3′ and 5′?CGTAGCCACGTGCTCTAATCCTC?3′ were used. PCR reaction mixtures contained 10 mM Tris?HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 200 μM of each dNTP, 0.4 μM of each primer, and 0.5 units Taq polymerase. The reactions were performed for 30 cycles under a regime of 50 s at 94°C, 40 s at 58°C, 1 min at 72°C. Except for the ITS gene locus, the other two (matK and rbcL) gene failed the QC for the amplification. Therefore, only the ITS was amplified and further analyzed by DNA sequencing to get the following data.
The sample was sent to the Eurofins Genomics lab in Bangalore India for sequencing done with the Sanger method. The results were than analyzed as per need with nBLAST tool of NCBI and other specific software for phylogenetic and sequence analysis based on Seq Scanner 2.0 software.
Results
Blast output
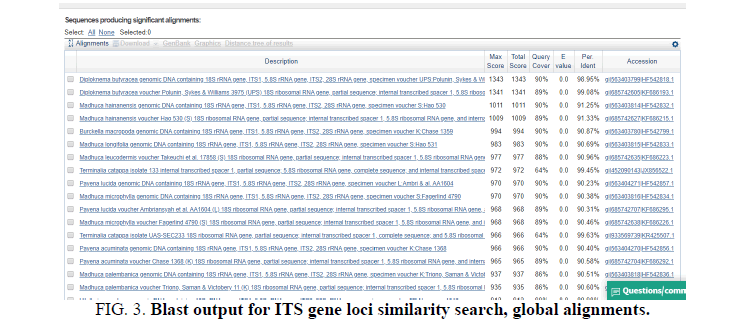
The Final sequence obtained of ITS region with its reverse and forward region was as follows (FIG. 3).
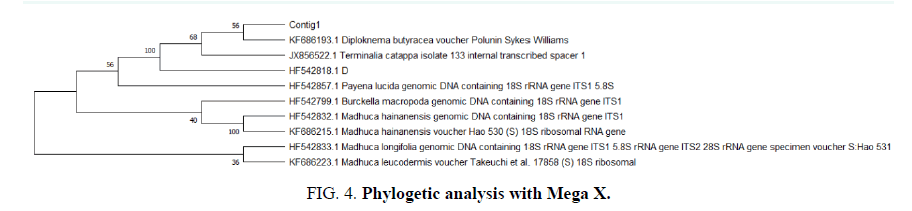
The Mega X Phylogenetic analysis of first 10 sequences for Cotig 1 of ITS gene loci with respect to BLAST output (FIG. 4).
Ancestral states were inferred using the Maximum likelihood method and Tamura-nei model. The tree shows a set of possible nucleotides (states) at each ancestral node based on their inferred likelihood at site 1. The initial tree was inferred using the method. The rates among sites were treated as being uniform among sites (Uniform rates option). This analysis involved 10 nucleotide sequences. There were a total of 1017 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
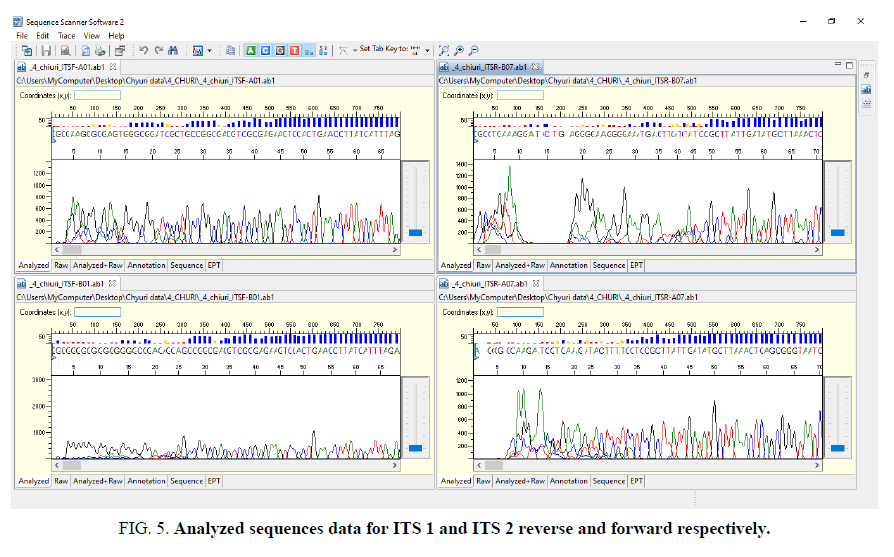
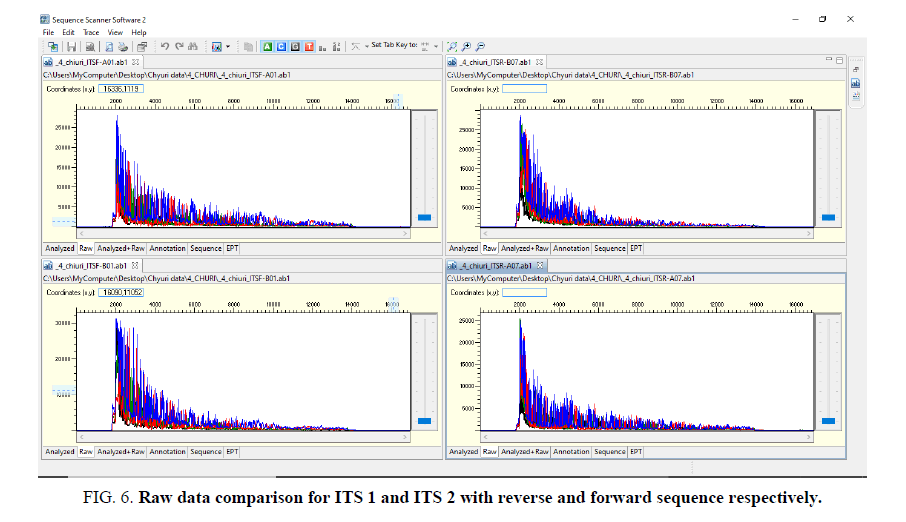
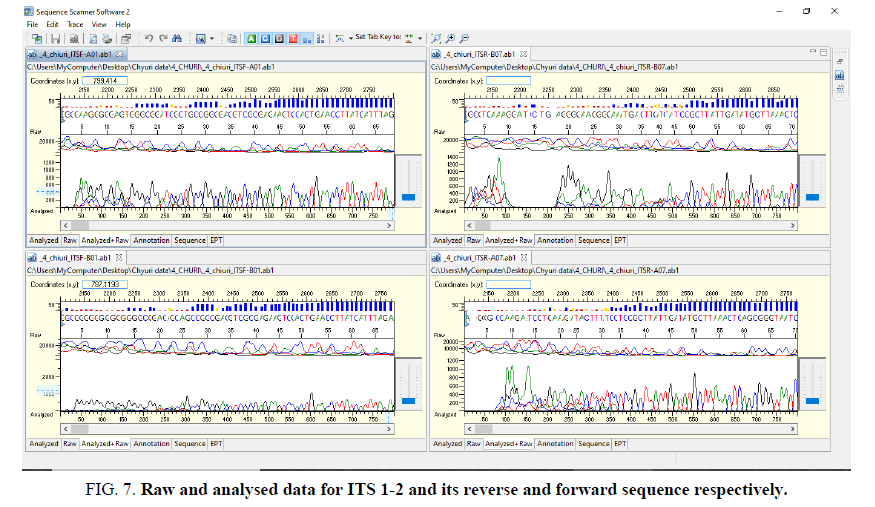
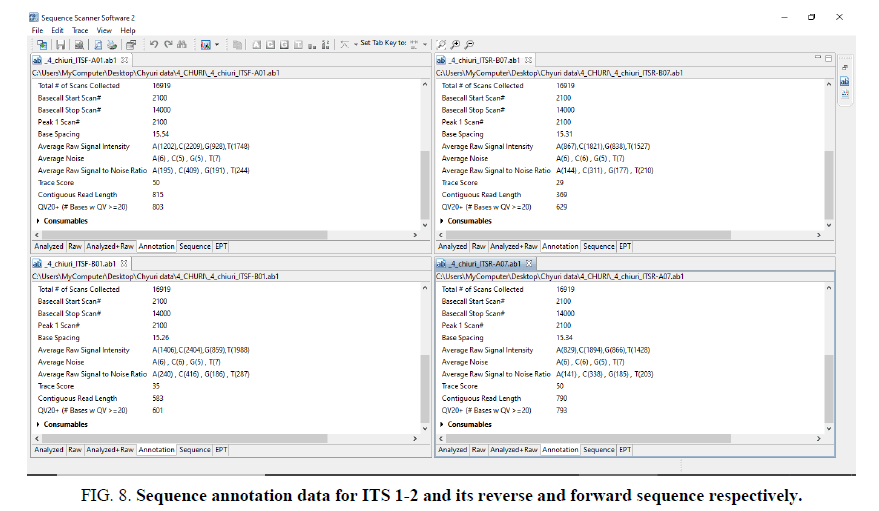
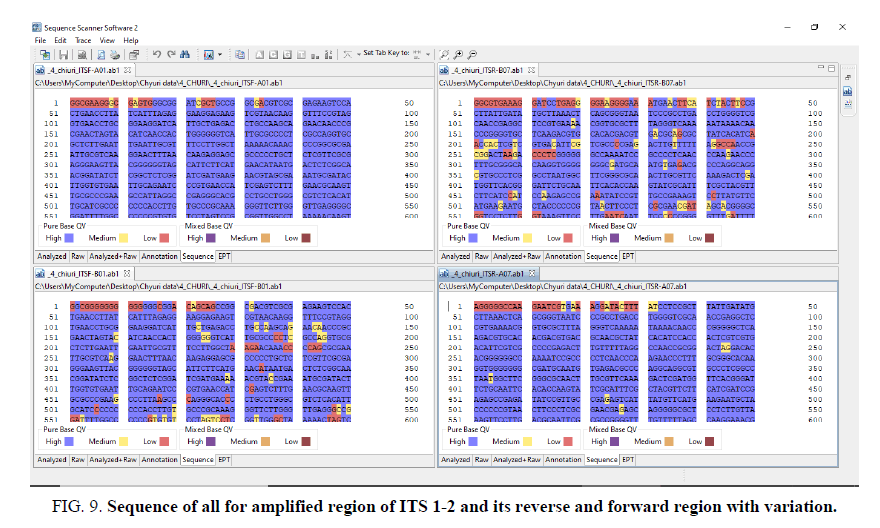
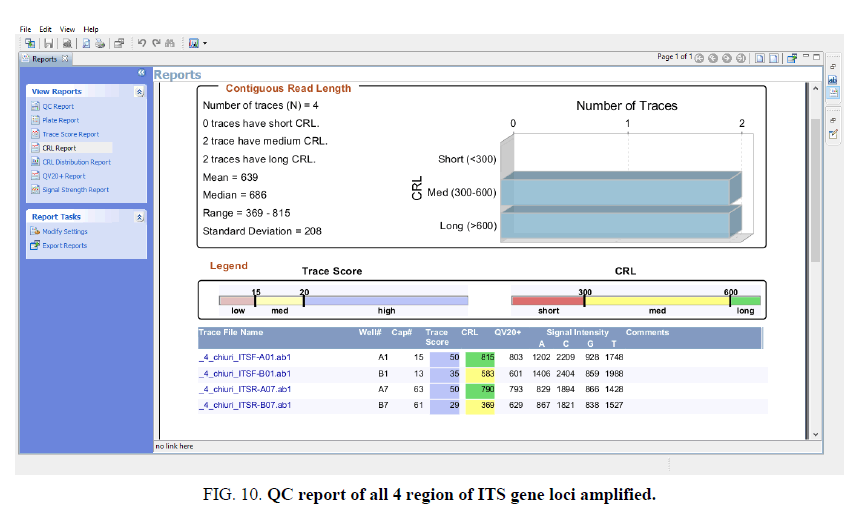
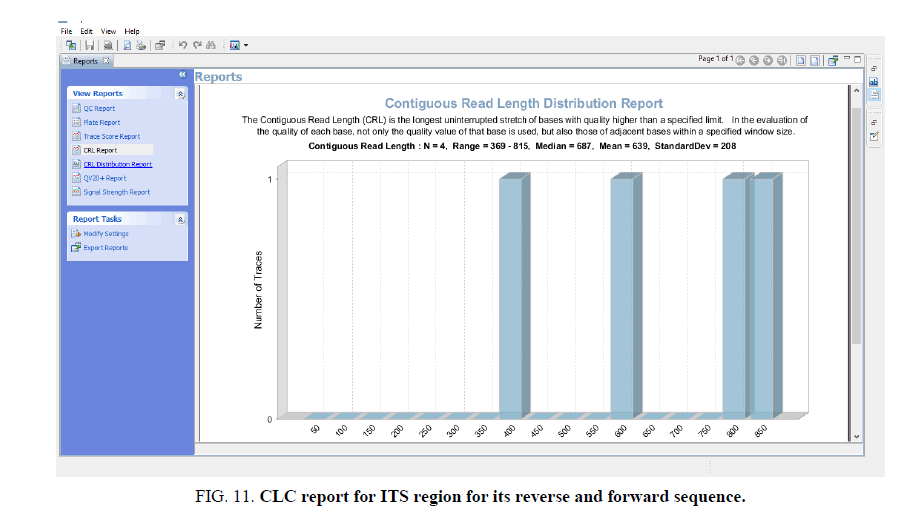
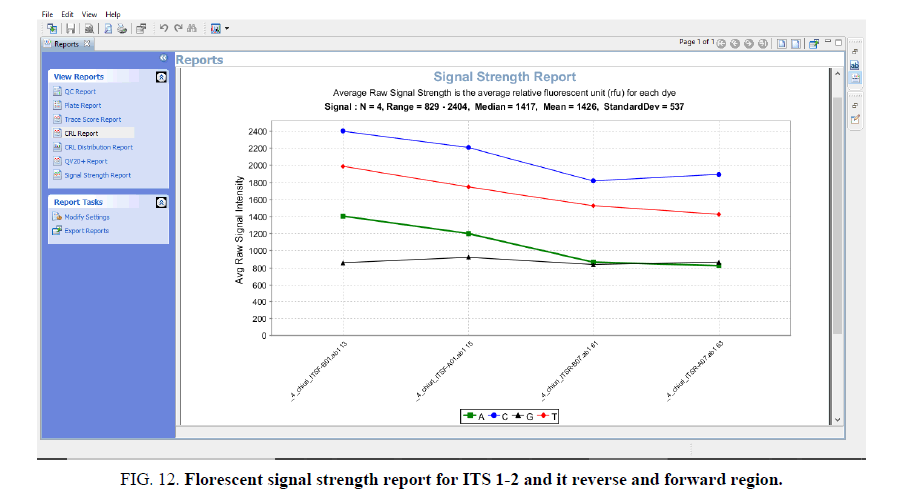
Sequence assembly and analysis report based of sequence scanner software in 6 different modules (FIG. 5-12).
Figure 9. Sequence of all for amplified region of ITS 1-2 and its reverse and forward region with variation.
The DNA barcoding was done using omics approach and phylogenetic tree shows the sequence similarity with Clustal 2.0 software as (FIG. 13).
Diplokhema butyracea spp. is a plant of Nepal that is widely used and abundantly available from 500 m to 2000 m altitudes from the eastern to the western part of the country. Community people use this plant for many purposes. The leaves are used as fodder for the cows and goats whereas the long aged tree logs are used for household furniture. The major parts of the plant used are its flowers and seeds. Flowers are used as vegetables and the pulp of ripe fruits can be used as butter for daily life. Due to the high content of pulp in each fruit, along with a high amount of fat, butter is a good business for local people with this plant as it is considered to be one of the best cash generating crops for people residing in the mountainous region of Nepal.
Data analysis by Mega X software using bootstrap value 500 using NJ method of likelihood it is found the genetic variation of this species shown in FIG. 3 and 4 which clearly indicate the species diversity with respect to the ITS gene loci only have potential barcode region which can be used it as an authentic marker, even more other barcode loci didn’t show the direct role in species diversification might be due to geological altitudinal variation or environmental cause.
The other FIG. 5-10 shows the variation at nucleotides which is concern with the diversity of the species.
Due to its high economic value the plant it is also important for research, including genetic authentication that may help to preserve it as the indigenous property of this country. With the use of our DNA barcoding sequence, now onward one can conduct more elaborate researches for sources of indigenous medicinal herbs and even for the conservation and cultivation of this type of commonly used wild plants species. Identification of adulterating components in the herbal drug formulations can also be identified by barcoding and will provide for better health care as well. DNA barcodes based on biological sequences in databases should be developed for all the targeted and highly valued specific medicinal plants species. This can be the one of the most important events for the future economic development of Nepal.
Conclusion
Diplokhema butyracea spp. (Chiuri) is one of the major plants of mountain area communities in Nepal with high economic value. The barcoding of this plant and deposition of the sequence in online database (accession no MT497981) will help in the identification and conservation of this species as one of the major indigenous plants of Nepal. Research towards finding of different chemical constituents present in the flowers, seeds and pulps of this valuable plant, along with their quantities and bioactivities, medical values and food values is essential but yet to be done. We suggest that the use of DNA barcoding can be integrated into a workflow during floristic studies and at national herbaria in the region. This could significantly increase the number of identified specimens and improve knowledge about species distributions in natural populations. ITS combines the highest resolving power for discriminating closely related species with a high PCR and sequencing success rate across a broad range of Diplokhema butyracea. Our research is first elaboration of the DNA barcoding sequence of this species and will open the new avenues in the field of molecular biotechnology and research in herbal medicine and its application for economic development in Nepal.
Acknowledgments
The Research work was financially supported by the University Grant Commission Nepal and PhD research grant support by Nepal Academy of Science and Technology (NAST) Nepal. We are also thankful to Eurofins Genomic Laboratory Bangalore for their kind support for DNA Sequencing and data analysis. We are also thankful to Tirtha Maiya Shrestha for the taxonomic identification of the samples.
Conflict of Interest
The authors declare that they have no conflict of interests.
References
- https://ethnobiology.org/sites/default/files/pdfs/JoE/23-2/Sacherer2003.pdf.
- Shrestha S, Pandey B, Mishra BP. Honey value chain Analysis-A case study of gahate Village, Lamjung District of Nepal. Int J Multidiscip Res Dev. 2017;4;107-112.
- Sharma D, Shrestha T, Rapal N, et al. Screening and bioactivity measurement of high altitude medicinal plants of Nepal. Vegetos-Int J Plant Res. 2016;29:7.
- Lamichhane J, Chhetri SB, Bhandari M, et al. Ethnopharmacological survey, Phytochemical screening and Antibacterial activity measurements of high altitude medicinal plants of Nepal: A bioprospecting approach. Indian J Tradit Med. 2014;13:496-507.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547-1549.
- Bhattarai K., Shrestha TM, Bajracharya R, et al. Biological activities of three different medicinal plants from Himalayan Region of Nepal. Nepal J Sci Technol. 2010;11:139-146.
- Lamichhane J, Bhattarai K, Shrivastava AK, et al. Chemical constituents of Rhododendron lepidotum from Nepal Himalayas. Chem Nat Compd. 2014;50:767-769.
- Devkota HP, Watanabe T, Malla KJ, et al. Studies on medicinal plant resources of the Himalayas: GC-MS analysis of seed fat of chyuri (Diploknema butyracea) from Nepal. Phcog J. 2012;4:42-44.
- Sengupta A, Choudhury SKR. Triglyceride composition of Madhuca butyraceae seed fat. J Am Oil Chem Soc. 1978;55:621-624.
- Techen N, Parveen I, Pan Z, et al. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014;25:103-110.
- Hebert PD, Cywinska A, Ball SL, et al. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313-321.
- Sharma D, Shrestha TM, Lamichhane J. DNA Barcoding of Myrica esculanta (Kafal), An indigenous, multipurpose and medicinal plant species from Nepal, by rbcL and matK Gene. Acta Sci Pharm Sci. 2020;4:11-18.
- https://webpages.uncc.edu/~jweller2/pages/BINF8350f2011/BINF8350_Readings/Doyle_plantDNAextractCTAB_1987.pdf.