Original Article
, Volume: 15( 2)1,3,5-Trisubstituted-1H-Pyrazole Derivatives as New Antimicrobial Agents: Synthesis, Characterization and Docking Study
- *Correspondence:
- Abdula AM, Chemistry Department, College of Science, Al-Mustansiriyah University, Baghdad, Iraq, Tel: 964-7808838128; E-mail: ahm.chem@yahoo.com
Received: March 29, 2017; Accepted: April 19, 2017; Published: April 25, 2017
Citation: Ismail AH, Abdula AM, Mohammed MT, et al. Al-Bayati. 1,3,5-Trisubstituted- 1H-Pyrazole Derivatives as New Antimicrobial Agents: Synthesis, Characterization and Docking Study. Int J Chem Sci. 2017;15(2):126.
Abstract
A four new 1,3,5-trisubstituted pyrazole derivatives (2a-d) were synthesized from the reaction of corresponding 1,3-diaryl-2-propene-1-one compounds (1a-d) with phenyl hydrazine in glacial acetic acid. The new pyrazole derivatives were screened for their antimicrobial activities against several bacterial species and candida albicans. Autodock approach was used to examination the interaction modes of substituted pyrazoles within the active pocket of GlcN-6-P enzyme.
Keywords
Pyrazole; Chalcone; Antimicrobia; Docking cell lines
Introduction
1H-pyrazoles are of special attention as they constitute an important class of natural and synthetic products, many of which exhibit versatile biological activities. Among them, 1,3,5-trisubstituted-1H-pyrazoles are exhibited antitumor [1], herbicide [2], antibacterial [3], Antifungal [4], hypoglycemic [5], antidepressant, analgesic [6] and anti-inflammatory [7] activities. Recently and due to the growing needed to find and seeking new antimicrobial agents we are focusing in this article into synthesis, characterization and antimicrobial study of novel 1,3,5-trisubstituted-1H-pyrazoles against several gram positive and gram negative bacterial species and also against Candida albicans. Docking approach was also achieved in order to examine the binding mode of the pyrazole derivatives inside the binding site of glucosamine-6-phosphate synthase (GlcN-6-P) which represents the efficacious target in antimicrobial chemotherapy [8].
Experimental
All the reaction inspected by TLC, starting materials and solvents were purchased from Sigma Al-drich company and used without purification. Melting points were specified on an electro-thermal capillary apparatus and are uncorrected; FT-IR measurements were recorded on a Shimadzu model FTIR-8400S. Mass spectra were re-corded on a Shimadzu GCMS-QP2010 Ultra apparatus. 1H-NMR spectra were gained with a Bruker spectrophotometer model ultra-shield at 300 MHz in DMSO-d6 solution with the TMS as internal standard. Note: in 1H-NMR spectra, the signals at δ 2.5 and 3.35 are for solvent (DMSO-d6) and dissolved water in DMSO-d6, respectively.
Synthesis
Synthesis of 1,3-diaryl-2-propene-1-one derivatives (1a-d): 1,3-diaryl-2-propene-1-one derivatives were prepared according to the modified procedure described in literature [9]. To a different methyl ketone (1 mmol) in ethanol (10 mL), sodium hydroxide (40%, 1 mL) was added and the mixture was stirred for ½ h. After that corresponding aromatic aldehyde (1 mmol), added and the reaction mixture was stirred overnight. The reaction mixture was allowed to stand at room temperature. The precipitated solid was dried and recrystallized from absolute ethanol.
(E)-1-(4-aminophenyl)-3-(thiophen-2-yl) prop-2-en-1-one (1a): Yellow powder, yield 96%, M.P.110°C to 113°C; IR (cm-1): 3427, 3313 (NH2), 3215 (aromatic C-H), 3099 (aliphatic C-H), 1631 (C=O), 1597 (CH=CH), 1575 (C=C). 1H-NMR (300 MHz, DMSO-d6) δ (ppm): 6.12 (s, 2H, NH2), 6.61 (d, 2H, Ar-H, j=8.56 Hz), 7.15-7.86 (m, 7H, 5 Ar-H, -CH=CH). GCMS (NCI) m/e: 229 M+ for C13H11NOS.
(E)-1-(4-hydroxyphenyl)-3-(thiophen-2-yl) prop-2-en-1-one (1b): Yellow powder, yield 65%, M.P.163°C to 165°C; IR (cm-1): 3200 broad peak (OH), 3101 (aromatic C-H), 3020 (aliphatic C-H), 1635 (C=O), 1602 (CH=CH), 1585 (C=C). 1H-NMR: (300MHz, DMSO-d6) δ (ppm): 6.88-6.90 (m, 2H, Ar-H), 7.11-7.17 (m, 1H, Ar-H), 7.52-8.02 (m, 6H, 4 Ar-H, CH=CH), 10.47 (s, 1H, OH). GCMS (NCI) m/e: 230 M+ for C13H10O2S.
(E)-1-(4-aminophenyl)-3-(3,4-dimethoxyphenyl) prop-2-en-1-one (1c): Yellow powder, yield 93%, M.P.122°C to 125°C; IR (cm-1): 3446, 3348 (NH2), 3242 (aromatic C-H), 2962 (aliphatic C-H), 1641 (C=O), 1597(CH=CH), 1579 (C=C). 1H-NMR (300MHz, DMSO-d6) δ (ppm): 3.80 (s, 3H, OCH3), 3.85 (s, 3H, OCH3), 6.10 (s, 2H, NH2), 6.60 -7.94 (m, 9H, 7 Ar-H, CH=CH). GCMS (NCI) m/e: 283 M+ for C17H17NO3.
(E)-3-(3,4-dimethoxyphenyl)-1-(4-hydroxyphenyl)-prop-2-en-1-one (1d): Yellow powder, yield 63%, M.P. 200°C to 202°C; IR (cm-1): 3238 broad peak (OH), 3072 (aromatic C-H), 2951 (aliphatic C-H), 1639 (C=O), 1597 (CH=CH), 1552 (C=C). 1H-NMR (300MHz, DMSO-d6) δ (ppm): 3.81(s, 3H, OCH3), 3.86 (s, 3H, OCH3), 6.90 (d, 2H, Ar-H, j=8.63 Hz), 7.04 (d, 1H, CH=CH, j=7.45Hz), 7.34-8.08 (m, 6H, 5 Ar-H, CH=CH), 10.39 (s, 1H, OH). GCMS (NCI) m/e: 284 M+ for C17H16O4.
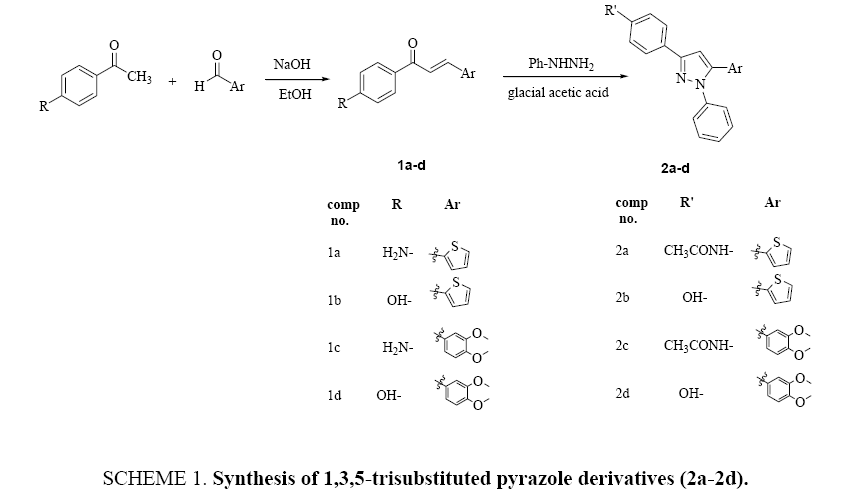
Synthesis of 1,3,5-trisubstituted pyrazoles (2a-2d): These compounds were synthesized according to modified procedure described in the reference [10]. To the mixture of corresponding chalcone 1a-1d (1 mmol) in 20 mL of glacial acetic, phenyl hydrazine (1 mmol) was added and the reaction mixture was refluxed overnight. After completion of reaction as monitored by TLC using ethyl acetate: hexane system (5:5 and 3:7), the reaction mixture was cooled and poured into ice water. The precipitate washed with cold water and recrystallized from ethanol.
N-(4-(1-phenyl-5-(thiophen-2-yl)-1H-pyrazol-3-yl)-phenyl)-acetamide (2a): Brown powder, yield 60%, M.P. 90°C to 93°C; IR (cm-1): 3275 (NH), 3103 (aromatic C-H), 2964 (aliphatic C-H), 1670 (C=O), 1651 (C=N), 1593 (C=C). 1H-NMR (300 MHz, DMSO-d6) δ (ppm): 2.06 (s, 3H, CH3), 6.60-8.15 (m, 13 H, 12 Ar-H, pyrazol-H), 10.05 (s, 1H, NH). GCMS (NCI) m/e: 359 M+ for C21H17N3OS.
4-(1-Phenyl-5-thiophen-2-yl-1H-pyrazol-3-yl)-phenol (2b): Brown powder, yield 82%, M.P. 62°C to 64°C; IR (cm-1): 3150 broad peak (OH), 3034 (aromatic C-H), 2950 (aliphatic C-H), 1641(C=N), 1597 (C=C). 1H-NMR (300 MHz, DMSO-d6) δ (ppm): 6.75-8.10 (m, 13 H, 12 Ar-H, pyrazol-H), 11.50 (s, 1H, OH). GCMS (NCI) m/e: 318 M+ for C19H14N2OS.
N-(4-(5-(3,4-dimethoxyphenyl)-1-phenyl-1H-pyrazol-3-yl)-phenyl)-acetamide (2c): Brown powder, yield 85%, M.P. 68-71°C; IR (cm-1): 3404 (NH), 3050 (aromatic C-H), 2929 (aliphatic C-H), 1672 broad peak (C=O, C=N), 1593 (C=C). 1H-NMR (300MHz, DMSO-d6) δ (ppm): 2.06 (s, 3H, CH3), 3.75 (s, 3H, OCH3), 3.81 (s, 3H, OCH3), 6.80-8.15 (m, 13H, 12 Ar-H, pyrazol-H), 10.15 (s, 1H, NH). GCMS (NCI) m/e: 413 M+ for C25H23N3O3.
4-(5-(3,4-dimethoxyphenyl)-1-phenyl-1H-pyrazol-3-yl)-phenol (2d): Brown powder, yield 34%, M.P 88°C to 91°C; IR (cm-1): 3281 broad peak (OH), 3037 (aromatic C-H), 2931(aliphatic C-H), 1651(C=N), 1597 (C=C). 1H-NMR (300 MHz, DMSO-d6) δ (ppm): 3.70 (s, 3H, OCH3), 3.75 (s, 3H, OCH3), 6.75-7.65 (m, 12H, Ar-H), 8.59 (s, 1H, pyrazol-H), 9.80 (s, 1H, OH). GCMS (NCI) m/e: 372 M+ for C23H20N2O3.
Antimicrobial study
The 1,3,5-trisubstituted pyrazole derivatives (2a-2c) were screened for their antimicrobial activity against Escherichia coli, Pseudomonas aeruginosa (gram positive), Staphylococcus aureus, Bacillus subtilis (gram positive) as well as candida albicans using the well diffusion method [11]. Dimethyl sulfoxide (DMSO) was used as a control and the test was achieved at 10 mg/mL and 100 mg/mL concentration using DMSO solvent. Each experiment was run in triplicate and the average reading was recorded.
Docking study
Auto Dock 4.2 bundle software was used to inspect the affinity of 1,3,5-trisubstituted pyrazole derivatives (2a-2d) to the binding site of GlcN-6-P synthase. The pdb file construction of enzyme as receptor was acquired from Protein Data Bank (PDB code 1 MOQ) and used as a static structure. Water molecules were extracted, natural inhibitor was removed and the missing hydrogen atoms were added to the protein residues. The two-dimensional structures of pyrazole derivatives (2a-2d) were drawn using Chem Draw ultra 7.0 and converted to mol formatted file using open Babel 2.3.1 software. During the docking, the grid of 62 A° in X, Y and Z were specified as 30.5, 17.5 and -2.2, respectively with points separated by 0.358 A° was built on the center of the catalytic site of enzyme. Lamarckian Genetic was utilized as the docking algorithm with 10 runs and 150 population size. As a default, the maximum number of energy evaluation equal to 2.5 × 105 and 27 × 103 generations were optimized [12, 13].
Results and Discussion
Synthesis
Chalcone derivatives (1a-d) were obtained by the reaction of methyl ketones with substituted aldehydes in ethanol in the presence of sodium hydroxide (Claisen-schmidt method) Scheme 1.
The 1,3-diaryl-2-propene-1-one derivatives (1a-1d) were characterized using IR, 1H-NMR and GCMS technique. The IR spectrum of (E)-1-(4-aminophenyl)-3-(thiophen-2-yl)-prop-2-en-1-one (1a) derivative shows absorption at 3427 and 3313 cm-1 related to -NH2 and 1631 cm-1 which is due to the stretching frequency of C=O group, while the aliphatic CH=CH and C=C stretching frequencies appeared at 1595 cm-1 and 1575 cm-1 respectively.
The 1H-NMR of 1a showed a singlet signal at δ 6.12 due to NH2 protones, doublet signals at δ 6.61 related to two aromatic protons, while the other aromatic protons and the COCH=CH protons appears as a multiplet signal at 7.15 ppm to 7.86 ppm. The GCMS peak at 229 further confirms the molecular ion M+ of compound. 1,3,5-Trisubstituted pyrazole derivatives (2a- 2d) were synthesized by refluxed of (1,3-disubstituted)-prop-2-ene-1-one (1a-d) and phenyl hydrazine mixture in glacial acetic acid as summarized in Scheme 1. The synthesized derivatives were characterized after recrystallization from an appropriate solvent by recording their IR, 1H-NMR and GCMS spectra. The IR spectrum of compound 2a showed absorption at 3275 referring to stretching frequency of N-H and 1670 cm-1 related to amide carbonyl (C=O), while absorptions of C=N and C=C groups appear at 1651 cm-1 and 1593 cm-1, respectively. The 1H-NMR of pyrazole derivative 2a showed singlet appeared at δ 2.06 due to methyl protons of amide group (COCH3) and multiplet signals at δ 6.60-8.15 related to aromatic protons as well as pyrazole proton. The NH amide proton appears as a singlet at 10.05 ppm. The molecular ion (359) M+ of pyrazole derivatives (2a) strongly confirmed by GCMS spectrum. Physical properties, spectral information, and mass analysis of all the synthesized compounds are illustrated in the experimental part.
Antimicrobial study
This research depicted the In Vitro assay of the 1,3,5-trisubstituted pyrazole derivatives (2a-c) against several microbial species (Table 1). The In Vitro assay achieved using two concentrations of synthesized derivatives 10 and 100 mg/mL. All the synthesized derivatives 2a-2d, especially compound 2b exhibited promising activities against candida albicans at 10 and 100 mg/ mL concentrations. The promising discovered pyrazole derivative 2d shows manifest antimicrobial activity against all bacterial species as well as candida albicans as described by Table 1.
| Pyrazole derivatives | Inhibition zone (mm) at 10 and 100 mg/mL | ||||
|---|---|---|---|---|---|
| Gram Negative | Gram Positive | Fungi | |||
| E.coli | P.aeruginosa | S. aureus | B.subitilis | C.albicans | |
| 2a | - | - | - | - | 15, (21)* |
| 2b | - | - | 15, (15)* | 15, (17)* | 16, (21)* |
| 2c | - | - | - | -, (14)* | 13, (17)* |
| 2d | -, (14)* | 19, (20)* | -, (11)* | 10, (12)* | 12, (18)* |
(-): No inhibition observed*: Inhibition zone in mm at 100 mg/mL
Table 1: Antimicrobial activity of 1,3,5-trisubstitutedpyrazole derivatives against versatile microbial species.
Docking study
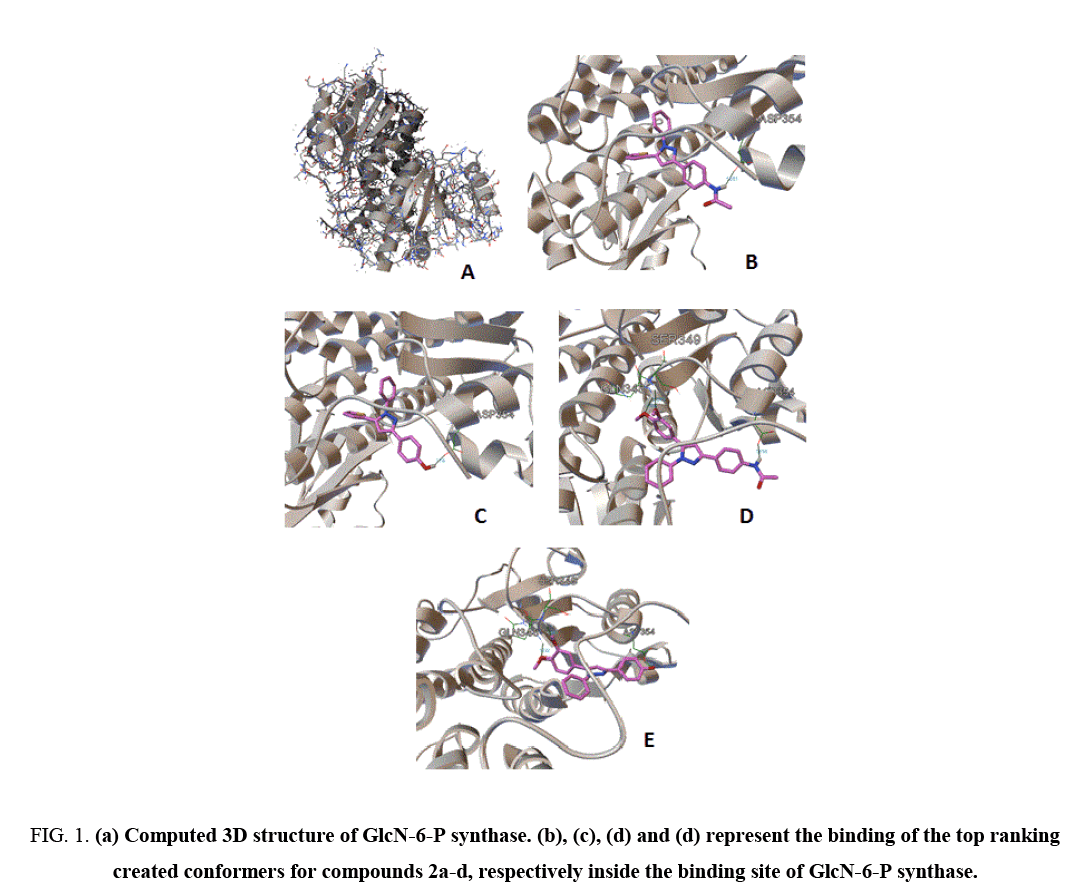
Docking approaches are computational methods for the inspection of the possible binding modes of a small molecule to a given receptor, enzyme or another binding site [13]. Docking study was achieved to explore the binding mode of the synthesized pyrazole derivatives (2a-2c) inside the binding cavity of glucosamine-6-phosphate synthase the potential target for antibacterial and antifungal agents. The binding site of GlcN-6-P enzyme included the following residues, Cys 300, Gly 301, Thr 302, Ser 303, Ser 347, Gln 348, Ser 349, Thr 352, Val 399, Ser 401, Ala 602 and Lys 603 as described by the X-ray crystallography [14]. Therefore, we used autodock 4.2 to evaluate the binding energy of ligands inside the known 3D structure of target enzyme. AutoDock 4.2 software compose of two major programs, autogrid that pre-calculates grid maps of interaction energies for different atom kinds of ligand with a macromolecule and autodock, which finalize the docking of the ligand to fixed grids [12]. For the typical systems, docking is achieved using a Lamarckian Genetic Algorithm (LGA), it is run divers times to give several docked conformations (ten conformers by default) classifying according to their binding and intermolecular energies. Different parameters were also portending by the autodock program such as inhibition constant, number of hydrogen bonds and others. The binding of the top generated conformers for pyrazole compounds 2a-2c inside the binding pocket of target enzyme illustrated by Figure 1, while Table 2 point out the molecular docking parameters of compounds 2a-2c.
| Pyrazole derivatives | Binding Energy (Kcal mol-1) |
Inhibition constant (µM) | Intermolecular energy (kcalmol-1) | H-bonds | Bonding | |
|---|---|---|---|---|---|---|
| 1 | 2a | -6.65 | 13.46 | -7.84 | 1 | LIG:H:ASP354:OD2 |
| 2 | -6.65 | 13.45 | -7.84 | 1 | LIG:H:ASP354:OD2 | |
| 3 | -6.64 | 13.50 | -7.84 | 1 | LIG:H:ASP354:OD2 | |
| 4 | -6.64 | 13.68 | -7.83 | 1 | LIG:H:ASP354:OD2 | |
| 5 | -6.64 | 13.67 | -7.83 | 1 | LIG:H:ASP354:OD2 | |
| 6 | -6.63 | 13.84 | -7.82 | 1 | LIG:H:ASP354:OD2 | |
| 7 | -6.63 | 13.69 | -7.83 | 1 | LIG:H:ASP354:OD2 | |
| 8 | -6.61 | 14.28 | -7.80 | 1 | LIG:H:ASP354:OD2 | |
| 9 | -6.60 | 14.42 | -7.80 | 1 | LIG:H:ASP354:OD2 | |
| 10 | -6.58 | 15.01 | -7.77 | 1 | LIG:H:ASP354:OD2 | |
| 1 | 2b | -6.46 | 18.52 | -7.65 | 1 | LIG:H:ASP354:OD2 |
| 2 | -6.45 | 18.66 | -7.64 | 1 | LIG:H:ASP354:OD2 | |
| 3 | -6.45 | 18.69 | -7.64 | 1 | LIG:H:ASP354:OD2 | |
| 4 | -6.45 | 18.79 | -7.64 | 1 | LIG:H:ASP354:OD2 | |
| 5 | -6.45 | 18.74 | -7.64 | 1 | LIG:H:ASP354:OD2 | |
| 6 | -6.44 | 18.91 | -7.64 | 1 | LIG:H:ASP354:OD2 | |
| 7 | -6.44 | 19.18 | -7.63 | 1 | LIG:H:ASP354:OD2 | |
| 8 | -6.43 | 19.31 | -7.62 | 1 | LIG:H:ASP354:OD2 | |
| 9 | -6.40 | 20.27 | -7.60 | 1 | LIG:H:ASP354:OD2 | |
| 10 | -6.18 | 29.56 | -7.37 | 1 | LIG:H:ASP354:OD2 | |
| 1 | 2c | -7.74 | 2.13 | -9.53 | 3 | GLN348:HN:LIG:O; SER349:HN:LIG:O; LIG:H:ASP354:OD2 |
| 2 | -7.64 | 2.49 | -9.43 | 3 | GLN348:HN:LIG:O; SER349:HN:LIG:O; LIG:H:ASP354:OD2 | |
| 3 | -7.49 | 3.23 | -9.28 | 3 | GLN348:HN:LIG:O; SER349:HN:LIG:O; LIG:H:ASP354:OD2 | |
| 4 | -7.49 | 3.26 | -9.28 | 3 | GLN348:HN:LIG:O; SER349:HN:LIG:O; LIG:H:ASP354:OD2 | |
| 5 | -7.26 | 4.74 | -9.05 | 2 | LIG:H:ASP354:OD2; SER349:HN:LIG:O | |
| 6 | -7.26 | 4.74 | -9.05 | 2 | LIG:H:ASP354:OD2; GLN348:HN:LIG:O | |
| 7 | -7.11 | 6.17 | -8.90 | 2 | LIG:HASN600:O; THR352:HG1:LIG:O | |
| 8 | -7.00 | 7.34 | -8.79 | 2 | LIG:H:ASN600:O; THR352:HG1:LIG:O | |
| 9 | -6.83 | 9.83 | -8.62 | 1 | THR352:HG1:LIG:O | |
| 10 | -6.79 | 10.57 | -8.58 | 2 | LIG:H:ASN600:O; THR352:HG1:LIG:O | |
| 1 | 2d | -7.67 | 2.38 | -9.46 | 3 | LIG:H:ASP354:OD2; SER349:HN:LIG:O; GLN348:HN:LIG:O |
| 2 | -7.59 | 2.74 | -9.38 | 3 | LIG:H:ASP354:OD2; SER349:HN:LIG:O; GLN348:HN:LIG:O | |
| 3 | -7.54 | 2.99 | -9.33 | 3 | LIG:H:ASP354:OD2; SER349:HN:LIG:O; GLN348:HN:LIG:O | |
| 4 | -7.30 | 4.44 | -9.09 | 2 | LIG:H:ASP354:OD2; GLN348:HN:LIG:O | |
| 5 | -6.91 | 8.67 | -8.70 | 2 | LIG:H:ASN600:O; THR352:HG1:LIG:O | |
| 6 | --6.87 | 9.17 | -8.66 | 2 | LIG:H:ASP354:OD2; GLN348:HN:LIG:O | |
| 7 | -6.81 | 10.14 | -8.60 | 1 | LIG:H:ASP354:OD2 | |
| 8 | -6.79 | 10.55 | -8.58 | 2 | LIG:H:ASP354:OD2; GLN348:HN:LIG:O | |
| 9 | -6.21 | 27.97 | -8.00 | 0 | - | |
| 10 | -6.02 | 38.83 | -7.81 | 2 | LIG:H:SER349:OG; SER349:HN:LIG:O | |
Table 2: Docking parameters for the synthesized derivatives (2a-2d).
Figure 1: (a) Computed 3D structure of GlcN-6-P synthase. (b), (c), (d) and (d) represent the binding of the top ranking created conformers for compounds 2a-d, respectively inside the binding site of GlcN-6-P synthase.
The binding energy of the generated conformers for the potent discovered pyrazole derivative 2d was between -7.67, for the best conformer and -6.02 kcal mol-1. The best generated conformers of 2d fits the binding pocket residues of enzyme with three hydrogen bonds. The residues were ASP354, SER349 and GLN348, respectively, as depicted by Table 2. The calculated inhibition constants for the best pyrazole derivatives 2d were between 2.38 μM and 38.83 μM (Table 2) strongly enhanced the in vitro assay of discovered hit derivative.
Conclusion
Four new 1,3,5-trisubsituted pyrazole derivatives were synthesized and characterized using IR, 1H-NMR and GCMS Technique. The pyrazole compounds (2a-d) were screened against several bacterial species as well as against candida albicans. Compound 2d exhibited promising antimicrobial activity against all species. Docking study was achieved to exploring the binding affinity of tested derivatives inside the binding site of GlcN-6-P enzyme which considering the targeting enzyme for antimicrobial activity. The docking results strongly enhanced the in-vitro assay.
Acknowledgement
The authors would like to thank, Al-Mustansiriyah University, Baghdad, Iraq for its support in the present work.
References
- Dias LRS, Salvador RRS. Pyrazole carbohydrazide derivatives of pharmaceutical interest. Pharmaceuticals. 2012;5:317-24.
- Furuta KN. Synthesis and herbicidal activity of 1, 5-diaryl pyrazole derivatives. Chem Pharm Bull. 1999;47:857-68.
- Kumar R, Arora J,Ruhil S, etal. Synthesis and antimicrobial studies of pyrimidine pyrazole heterocycles. AdvChem. 2014;pp:12.
- Priyadarsini P, Ujwala B, Rao CV, et al. Synthesis and antimicrobial activity of some novel pyrazoles. Der Pharm lett. 2012;4:1123-8.
- Wright JB,Dulin WE,Markillie JH. The antidiabetic activity of 3,5-dimethylpyrazoles. J Med Chem. 1964;7:102-5.
- Baily DM,Geraffen, Patent. Chem Abstr. 1978;pp:89.
- Nargund LVG,Hariprasad V, Reddy GRN. Synthesis and anti-inflammatory activity of fluorinated phenyl styryl ketones and N-phenyl-5-substituted aryl-3-p-(fluorophenyl) pyrazolins and pyrazoles. J Pharm Sci. 1992;81:892-4.
- Bearne SL,Blouin C. Inhibition of Escherichia coli glucosamine-6-phosphate synthase by reactive intermediate analogues.JBiol Chem. 2000;275:135-40.
- Abdula AM. Synthesis, characterization and antibacterial activity of (E)-Ehalcone derivatives. Eur J Chem. 2013;4:207-10.
- Kumar R, Arora J,Ruhil S, et al. Synthesis and antimicrobial studies of pyrimidine pyrazole heterocycles. AdvChem. 2014;pp:1-12.
- Tomi IHR, Al-Daraji AHR, Abdula AM, et al. Synthesis, antimicrobial and docking study of three novel 2,4,5-triarylimidazole derivatives. J Saudi Chem Soc. 2016;20:S509-S16.
- Morris GM,Goodsell DS, Olson J. AutoDock 2.4 User Guide. 1996.
- Morris GM,Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. JComput Chem. 1998;9:639-62.
- Teplyakov A,Obmolova G,Badet-Denisot MA, et al. Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: Evidence from a 16 A? crystal structure of the isomerase domain. Structure. 1998;6:1047-55.